From IRBMReviewed by Olivia Frost
This article and associated images are based on a poster originally authored by Alaimo N., Fata F., Di Marino S., Carucci F., Ferrari F., Ventre D., Palombo S., Movetti S., Cerretani M., Benoni R., Berti F., Missineo A., Turcano L., Piai A., di Pasquale P., Malancona S., Montalbetti C., Carli I., Toniatti C. and Alli C. and presented at ELRIG Drug Discovery 2025 in affiliation with IRBM S.p.A.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
The COVID-19 pandemic has highlighted the need for new antiviral drugs with broad efficacy against coronavirus infections, likely able to prevent future outbreaks. The NSP13 helicase of SARS-CoV-2 is a crucial enzyme for viral replication, ensuring efficient viral RNA synthesis, replication, and transcription.
Due to its function and high conservation among coronaviruses, NSP13 is a promising therapeutic target for the treatment of Coronaviridae infections. Since helicases are challenging targets, multiple approaches were considered to increase the likelihood of identifying novel chemical matter for NSP13.
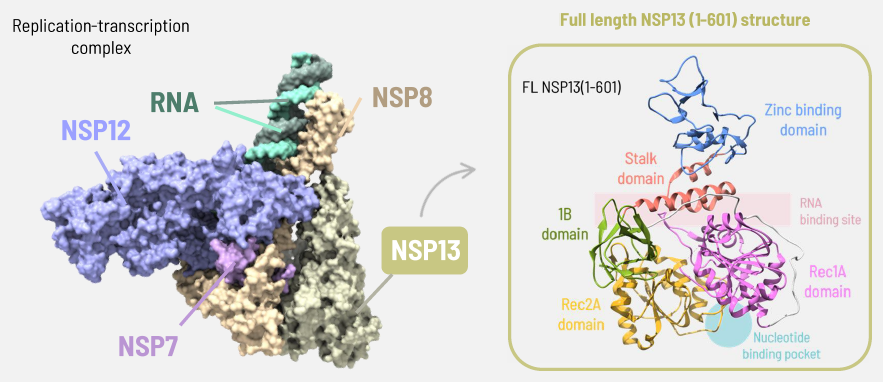
SARS-CoV-2 replication complex and NSP13 domain structure (PDB entry ID: 6XEZ and 6ZSL). Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
NSP13 helicase
- Zinc binding domain: Zinc-binding, with the aim of facilitating nucleic acid recognition
- Stalk domain: Linker between ZBD and domain 1B
- Rec1A domain contains conserved motifs for ATP binding/hydrolysis
- Rec2A domain cooperates with 1A for ATPase and unwinding activity
- 1B domain plays a role in modulating helicase activity.
Fragments screening by NMR
Unlike traditional high-throughput screening, Fragment Based Drug Discovery (FBDD) relies on identifying low-molecular-weight fragments that can be subsequently translated through rational design and medicinal chemistry into more potent and selective compounds
NSP13 protein production
The protein expression and purification protocol was adapted from Jia et al. and White et al. and optimized to produce high-quality samples.
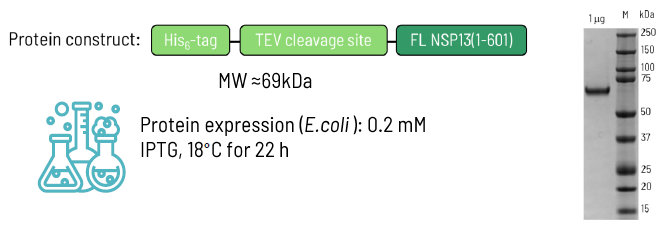
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
SDS-PAGE analysis
- Purity ≥90 %
- Yield: ~0.5 mg/L
- Concentration: ~20 μM
- Formulation buffer: PBS (pH 7.4), 100 μM DTT
Ligand-observed NMR experiments
Saturation Transfer Difference (STD), Water Ligand-Observed by Gradient SpectroscopY (WaterLOGSY), relaxation-based experiments (T2 and T1p) and DOSY (Diffusion-Ordered SpectroscopY) are well-established NMR methodologies for screening and hit optimization in fragment-based drug discovery (FBDD).
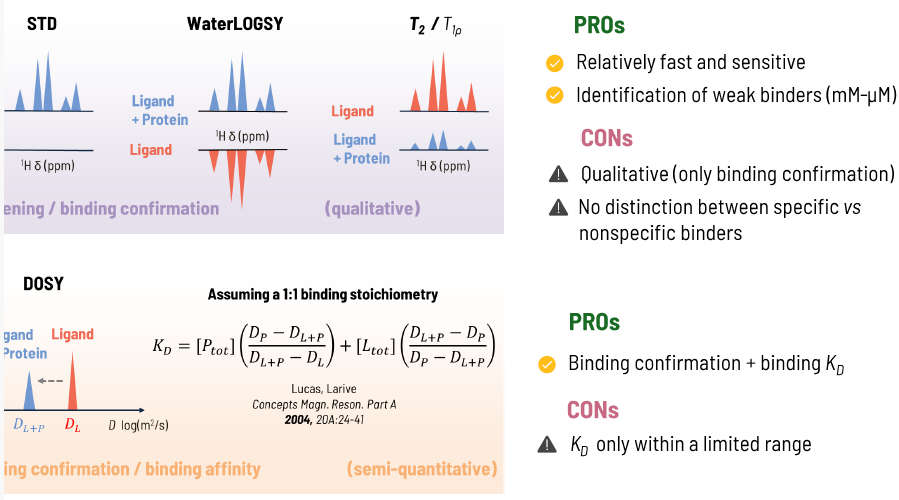
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
- STD: In the presence of a protein-ligand interaction, the saturation applied to the protein is transferred to the ligand, resulting in an increase in STD signal intensity for the ligand protons involved in binding
- WaterLOGSY: During protein-ligand binding, the presence of the protein induces polarization in surrounding water molecules, which is transferred to the ligand, allowing both bound and free molecules to be distinguished by opposite signals
- T2/T1p: Ligand relaxation rates increase upon binding to the protein, due to the larger molecular weight of the complex, leading to signal broadening
- DOSY: The diffusion coefficient of the ligand is measured and decreases upon binding to the protein. This change is proportional to the bound fraction of the ligand and can be correlated with its binding affinity (KD).
Screening results
An NMR-based fragment screening was performed using ~500 fragments from the IRBM collection, grouped into cocktails of 10 fragments each. ATP-γ-S was used as a positive control for assay setup and screening QC.
Generation of fragment mixtures
- 48 mixture cocktails of 10 fragments (467 fragments + ATP-γ-S)
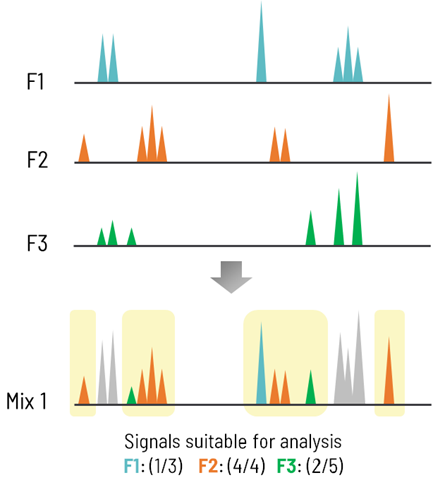
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
Experimental procedures
- Ligand QC: NMR
Protein QC: SEC, SDS-PAGE, binding to A TP-γ-S
- Acquisition of reference (fragment cocktail) and binding (fragment cocktail + NSP13) spectra to allow rigorous side-by-side comparison
- NMR experiments and data analysis:
Source: ELRIG (UK) Ltd.
| Experiment |
Analysis |
| STD |
+ |
(+) |
- |
| WaterLOGSY |
+ |
(+) |
- |
| T2 |
Avg. ΔT2 (%) |
+ positive response for all signals
(+) positive response for some signal
- negative response for all signals
Results:
The synergistic use of STD, WaterLOGSY, and T2 experiments led to the identification of 40 high-confidence fragment hits.
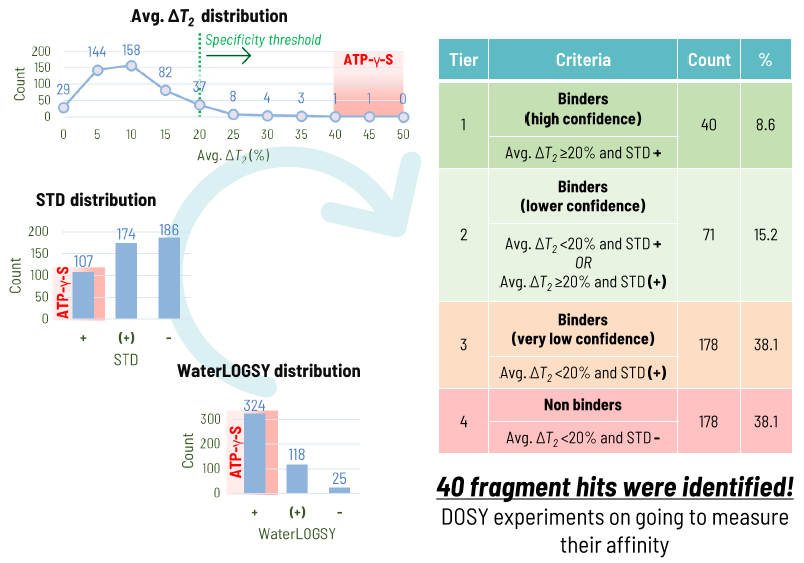
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
Hit confirmation and optimization
In parallel to DOSY NMR experiments, two different binding assays to NSP13 were implemented using Affinity Selection Mass Spectrometry (ASMS) and Surface Plasmon Resonance (SPR) techniques. These methods served as orthogonal readouts, for hit confirmation and optimization to support SAR.
ASMS
Compounds were tested on the NSP13 helicase at a fixed concentration and two time points. Molecules showing a response ratio greater than three were selected as binders.
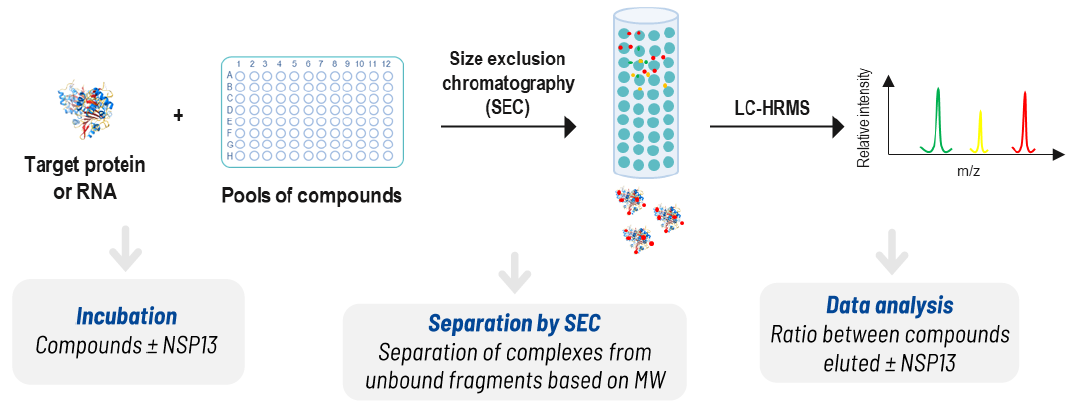
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
The best compounds were analyzed in a dose/ response experiment for KD determination.
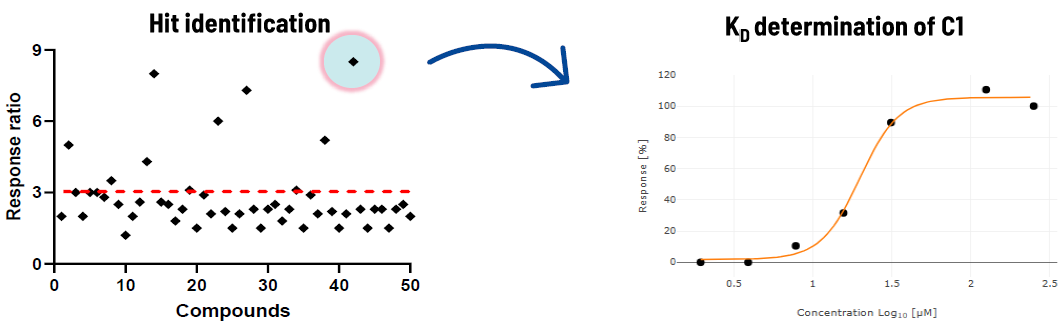
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
SPR
- AMP-NP is a non-hydrolyzable ATP analog that was used as an analyte to set up the SPR binding assay
- AMP-NP testing was repeated 12 times to assess protein stability on the chip (18 hours).
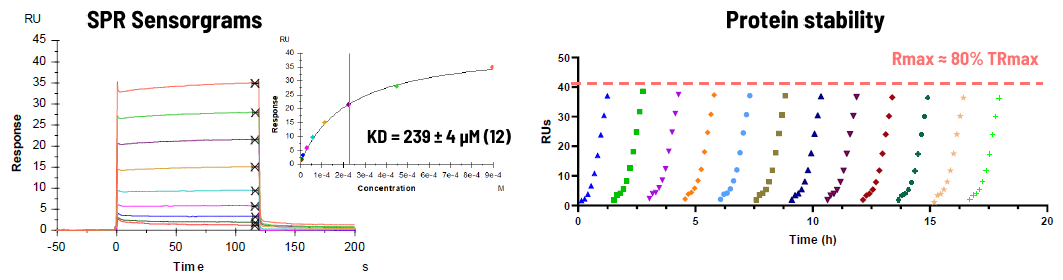
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
NSP13 showed high micromolar affinity to AMP-NP, and the protein remained stable up to 18 hours at 12 °C on the chip.
- The assay was applied to further validate compounds from ASMS and NMR
- Compounds C1 showed a KD in the low micromolar range.
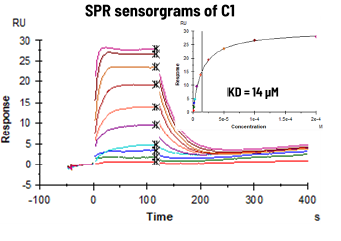
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
Biochemical assay
With the aim of identifying and characterizing NSP13 inhibitors, two biochemical assays were established: the ATPase and helicase unwinding assays. Both assays were used to screen the team's compound collection.
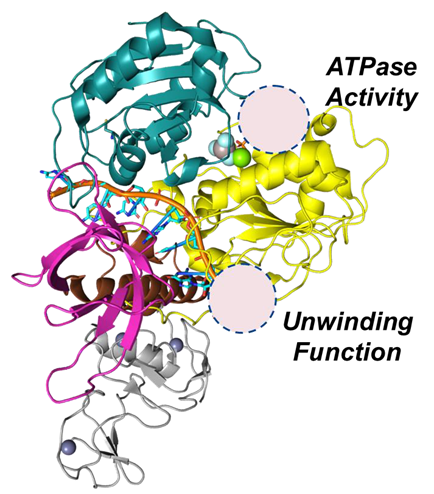
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
ATPase assay: The ATPase activity was monitored using ADPglo technology, and the amount of ATP consumption was measured.
FRET-helicase assay: The unwinding activity of NSP13 helicase was detected by monitoring the displacement of the quenched strand by the compounds (see scheme below).
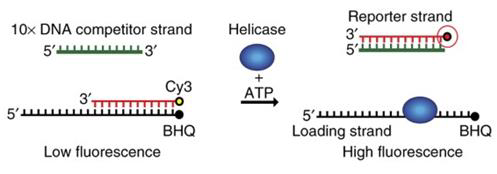
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
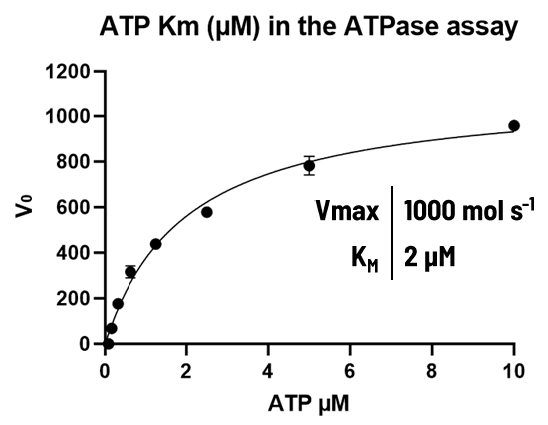
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
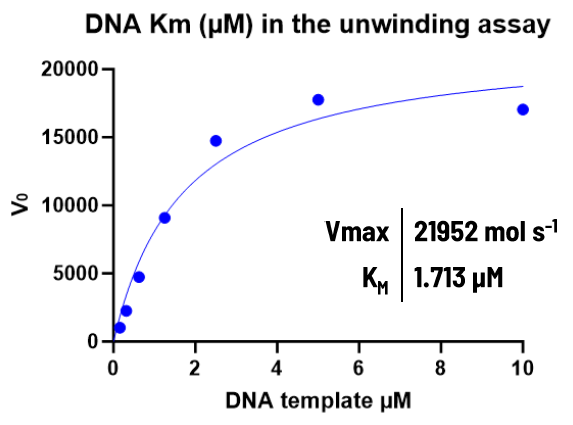
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
Hit ID screening funnel
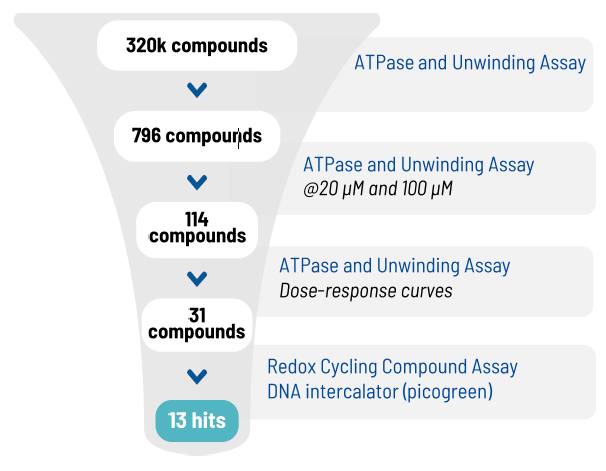
Image Credit: Image courtesy of Alaimo N., in partnership with ELRIG (UK) Ltd.
Even with a low hit rate, the campaign led to the identification of a small number of hits, which were further validated.
Conclusions
In this study, the research team reported the development of biochemical and biophysical assays aimed at identifying and characterizing inhibitors of the SARS-CoV-2 NSP13. A fragment-based screening campaign was conducted using NMR spectroscopy. As orthogonal readouts, ASMS, DOSY, and SPR were set up and validated.
Both the biochemical and fragment screening approaches led to the identification of novel chemical entities. The assays are now actively supporting structure-activity relationship (SAR) studies and facilitating the progression of the program from hit identification to lead optimization.
References
- Jia, Z., et al. (2019). Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Research, 47(12), pp.6538–6550. https://doi.org/10.1093/nar/gkz409.
- White, M.A., Lin, W. and Cheng, X. (2020). Discovery of COVID-19 Inhibitors Targeting the SARS-CoV-2 Nsp13 Helicase. The Journal of Physical Chemistry Letters, 11(21), pp.9144–9151. https://doi.org/10.1021/acs.jpclett.0c02421.
- Spratt, A.N., et al. (2021). Coronavirus helicases: attractive and unique targets of antiviral drug-development and therapeutic patents. Expert Opinion on Therapeutic Patents, 31(4), pp.339–350. https://doi.org/10.1080/13543776.2021.1884224.
About IRBM
IRBM is a leading contract research organization (CRO) based in Pomezia, near Rome, Italy.
The company specialises in integrated early-stage drug discovery across small molecules, peptides, and antibodies, providing end-to-end support from target validation and screening through to candidate nomination.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to consistently ensure the highest quality of content, making it readily accessible to all, and to maintain an inclusive organization that serves a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 13, 2025