This article and associated images are based on a poster originally authored by Ashleigh Hill, Marie-Louise Francis, Viktoria Brachmaier, Egle Vaitone, Rachel Forfar and Sandra Kuemper and presented at ELRIG Drug Discovery 2025 in affiliation with LifeArc.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Background:
The ubiquitin-proteasome system (UPS):
The UPS is necessary for biological degradation of damaged, misfolded, or unnecessary proteins; E1, E2, and E3 enzymes sequentially catalyse transfer of Ubiquitin (Ub) onto the lysine residues of substrates (fig. 1).
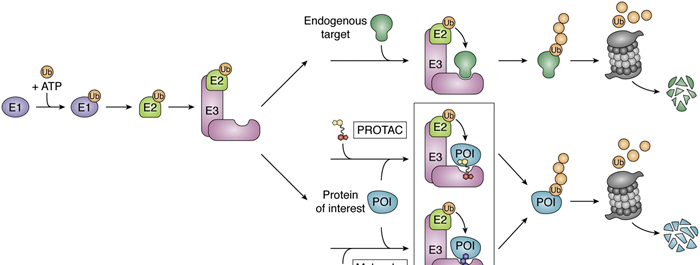
Figure 1. The UPS and TPD using PROTACs.1 Image Credit: Image courtesy of Ashleigh Hill et al., in partnership with ELRIG (UK) Ltd.
Targeted protein degradation (TPD):
TPD exploits the UPS to degrade disease-specific proteins. PROTACs (fig. 2) are heterobifunctional small molecules that hijack E3 ligases by forcing them into proximity with a protein of interest (POI), for its subsequent degradation via the proteasome (fig. 1). Currently exploited E3s are CRBN and VHL, largely confined to treatment of adult tumors.
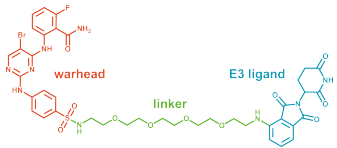
Figure 2. PROTAC (Proteolysis Targeting Chimera) structure.2 Image Credit: Image courtesy of Ashleigh Hill et al., in partnership with ELRIG (UK) Ltd.
Aims:
To identify and validate pediatric E3 ligases for TPD applications in solid childhood tumors.
Screening:
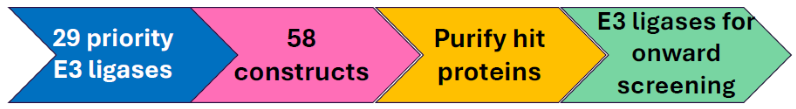
Image Credit: Image courtesy of Ashleigh Hill et al., in partnership with ELRIG (UK) Ltd.
- Developed and optimized a screening assay to investigate the degradation profiles of prioritized E3 ligases
- 29 priority E3 ligases were selected and filtered for:
- Tissue relevance
- Expression levels
- To ensure clinically relevant and reproducible results, two targets of interest and two cell lines were screened
Targets:
- FAK – Focal adhesion kinase
- PTK2B/ Pyk2 – Protein Tyrosine Kinase 2 Beta
Cell lines:
- A549
- U2OS- Pediatric
Workflow:
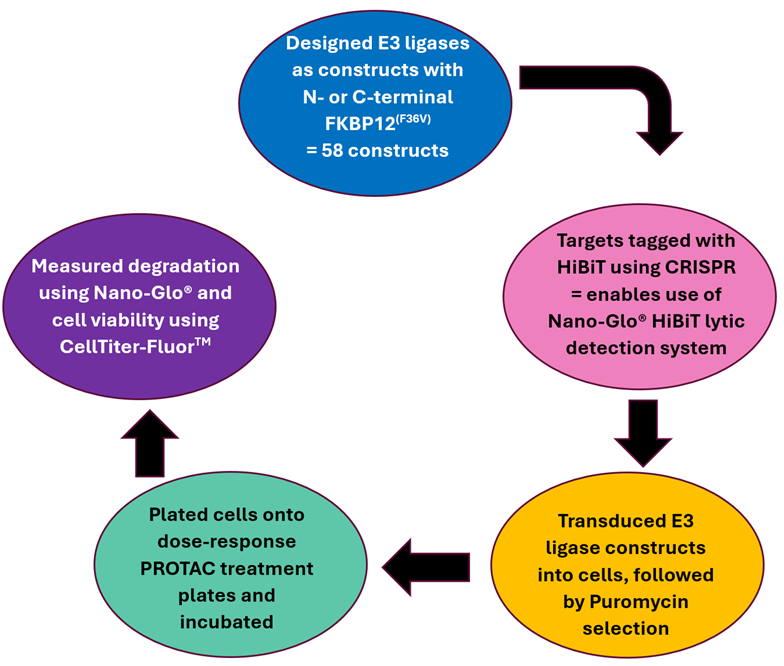
Image Credit: Image courtesy of Ashleigh Hill et al., in partnership with ELRIG (UK) Ltd.
Initial screen results - FAK (A549 cells):
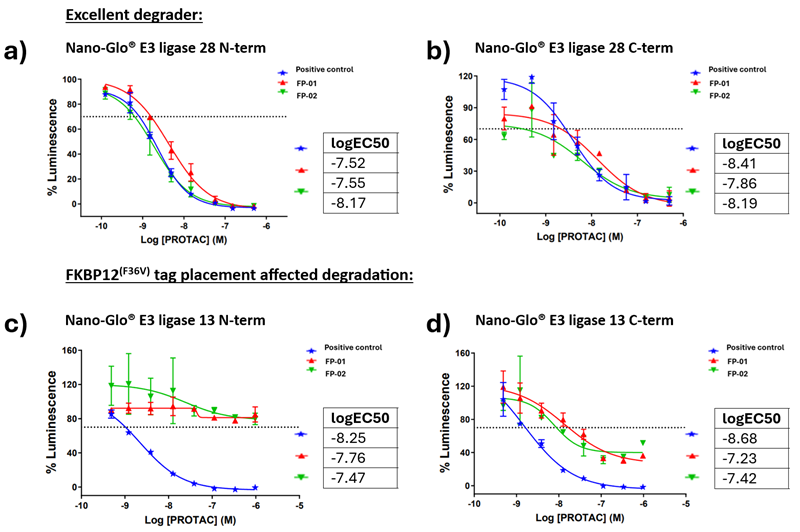
Figure 4. FAK degradation. FP – FAK PROTAC a) and b) show excellent degradation with both N- and C-terminal tagging of frontrunner E3 ligase 28. c) and d) Show C-terminal tag placement resulted in poor degradation compared with average degradation for N-terminal tag placement for E3 ligase 13.4 Image Credit: Image courtesy of Ashleigh Hill et al., in partnership with ELRIG (UK) Ltd.
- Limited cytotoxicity
- 24/ 29 Successful E3 ligases
- 15 “Excellent” Degraders
- DC50 < 1 μM
- Dmax > 30 % degradation
- Tag placement affected degradation
Follow-up screening (PTK2B & U2OS):
- Promiscuous PROTACs selected against control CRBN and VHL constructs. “Promiscuous” refers to the PROTAC warhead being a generic kinase binder
- 8/24 E3 ligases which degraded FAK also degraded PTK2B (A549)
- FAK degradation was not cell-line dependent –FAK degradation was repeatable in clinically relevant pediatric U2OS cells
Mechanism-of-action (MOA) confirmation:
- To confirm degradation via the proteasome, cells were incubated with:
a) 0.5 % DMSO, b) 1 μM Epoxomicin proteasome inhibitor, c) 1 μM MLN4924 neddylation inhibitor.
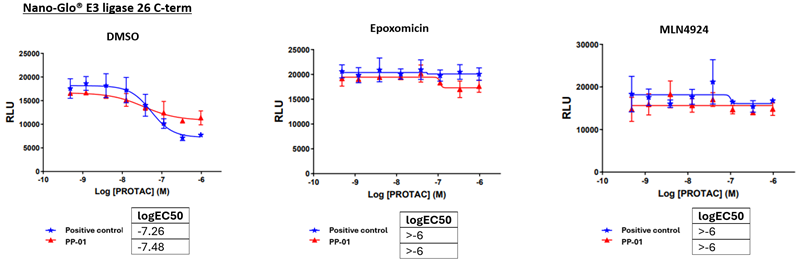
Figure 5. FAK degradation. PP – Promiscuous PROTAC a) Normal degradation with DMSO vehicle control, b) Proteasomal degradation confirmed by Epoxomicin, c) MLN4924 treatment confirms neddylation is required by Cullin-RING E3 ligase 26 for degradation.4 Image Credit: Image courtesy of Ashleigh Hill et al., in partnership with ELRIG (UK) Ltd.
Conclusions:
- Successfully developed and optimized a screening assay
- 15 novel E3 ligases showed excellent FAK degradation; six showed excellent PTK2B degradation
- FKBP12(F36V) tag placement is an important consideration in construct design
- Selected hits for protein purification
Next steps:
Complete protein purification for crystallisation of the hits and select E3 ligases for onward screening efforts.
References:
- Alabi, S.B. and Crews, C.M. (2021). Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. The Journal of Biological Chemistry, (online) 296, p.100647. DOI: 10.1016/j.jbc.2021.100647. https://www.jbc.org/article/S0021-9258(21)00433-6/fulltext.
- Nori, D., Coley, C.W., and Mercado, R. (2022). De novo PROTAC design using graph-based deep generative models. (online) arXiv. Available at: https://arxiv.org/abs/2211.02660.
- Promega. 2025.
- Analysis and graphs generated in GraphPad Prism version 9.4.1.
Acknowledgments:
Thank you to Marie-Louise Francis, Sarah Conway, Chloe Sanders, Chris Coward, Joao Pisco, Andy Merritt, the forced proximity assay team - Rachel Forfar, Sandra Kuemper, Viktoria Brachmaier, and the wider Cancer Grand Challenge: PROTECT Team.
About LifeArc
LifeArc® is a medical research charity making life science life-changing, transforming promising life science ideas into medical breakthroughs that change patients’ lives. We are self-funding and specialize in early-stage translation, advancing lab-based scientific discoveries to a point at which they can be developed into the next generation of diagnostics, treatments, and cures. We have been doing this for more than 25 years, and our work has resulted in a diagnostic for antibiotic resistance and four licensed medicines. This includes Keytruda®(cancer), Actemra® (rheumatoid arthritis), Tysabr® (multiple sclerosis), Entyvio® (Crohn’s disease), and a test for antimicrobial resistance.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free of charge to attend!
Our values
Our values are to always ensure the highest quality of content, that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate, and collaborate on an open-access basis. We achieve this through the provision of world-class conferences, networking events, webinars, and digital content.
Sponsored Content Policy: News-Medical.Net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.
Last Updated: Dec 18, 2025