From IRBMReviewed by Olivia Frost
This article and associated images are based on a poster originally authored by Cristina Alli, Chiara Soldati, Ottavia Cecchetti, Monica Bisbocci, Sara Pugliese, Mauro Cerretani, Elisa Beghetto, Marta Zavattieri, Ilaria Carli, Giacomo Paonessa, Rita Graziani, Libera Sessa, Giulia Di Marcantonio, Maria Vittoria Salvati, Alessio Sferrazza, Christian Montalbetti, Valentina Fodale and Carlo Toniatti and presented at ELRIG Drug Discovery 2025 in affiliation with IRBM S.p.A.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
Coronaviruses are known for their potential to cause global epidemics and pandemics, as demonstrated by the ongoing outbreak of SARS-CoV-2. This pandemic has presented significant health and economic challenges, underscoring the need for effective pan-coronavirus antiviral strategies.
Replication of the coronavirus genome requires continuous RNA synthesis, whereas transcription is a discontinuous process, which is characteristic of all coronaviruses and crucial for generating the subgenomic mRNAs required for producing viral proteins and successful viral replication.
Therefore, the discontinuous transcription process represents an interesting target for the development of pan-coronavirus inhibitors. To identify novel inhibitors of discontinuous transcription, the team developed a stable cellular model of Coronavirus replication, containing an SARS-CoV-based RNA replicon that lacks the viral genes encoding structural proteins and expresses NanoLuc Luciferase as a reporter gene.
In this poster, the team describes the development and validation of a high-throughput assay, and its use in a phenotypic screening campaign conducted using the IRBM's compound collection as well as an exhaustive panel of follow-up assays to enable the in-depth characterization of hits and the precise analysis of their mode of action.
Cellular model of coronavirus replication
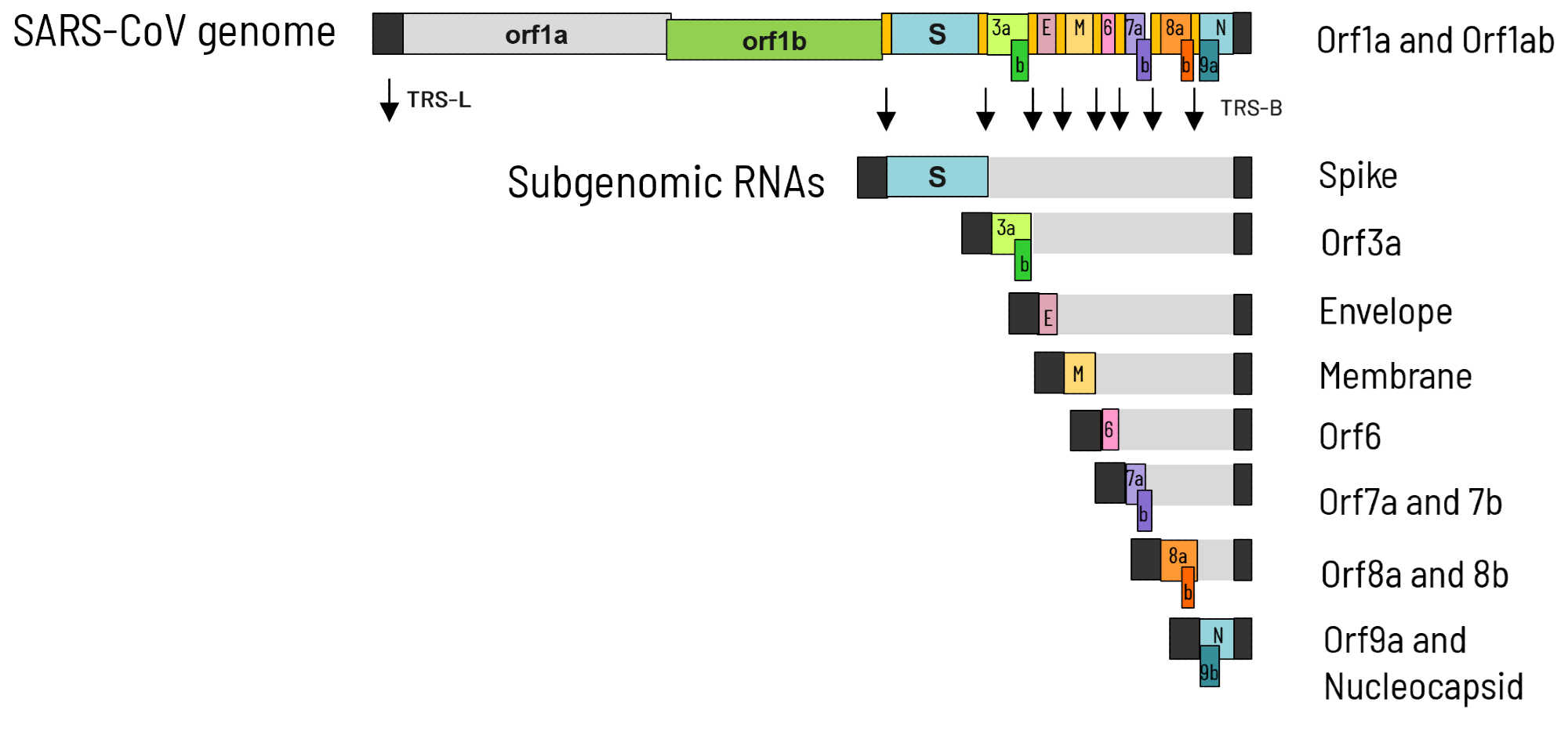
Fig.1. Coronavirus Discontinuous Transcription. Image Credit: Image courtesy of Malone et al., 2022, in partnership with ELRIG (UK) Ltd.
The SARS-CoV genome, like all coronavirus genomes, consists of a single-stranded positive-sense RNA molecule of about 30 kb encoding two overlapping polyproteins, ORF1a and ORF1ab.
After translation, these two polyproteins are processed into 16 mature proteins (NSP1 to NSP16) by the two virus-specific proteinases NSP3 and NSP5. The association of these mature proteins forms the replication-transcription complex (RTC), which mediates the synthesis of viral genomic RNA, gRNA (replication), and subgenomic mRNAs, sgRNA (transcription).
The synthesis of these two viral RNA species depends on specific cis-acting Transcription Regulatory Sequence (TRS) elements distributed along the viral genome. The sgRNAs encode all the structural proteins indispensable for viral particle formation.
Fig. 2. Replicon Generation by Gibson Assembly. Image Credit: Image courtesy of Cristina Alli et al., in partnership with ELRIG (UK) Ltd.
The replicon used in this work, SA062, was derived from the SARS-CoV isolate Sin2774 (AY283798). ORF1ab and all the accessory genes (ORF3a and 3b, ORF6, ORF7a and 7b, and ORF8) were retained.
At the same time, the structural proteins encoding sequences, Spike (S), Matrix (M), and Envelope (E), were substituted with two reporter genes, the NanoLuc Luciferase (Nluc) along with the Neomycin resistance, and the superfolded GFP (sfGFP).
The resulting sub-genome conserved all the RTC encoding sequences and the cis-acting elements responsible for the gRNA replication and sgRNA transcription, and thus can self-replicate. The N sequence, as part of the sfGFP sgRNA, was retained among the structural genes because of its demonstrated importance in protecting and launching the viral RNA synthesis.
Importantly, the expression of both the reporter genes depends on the discontinuous transcription process.
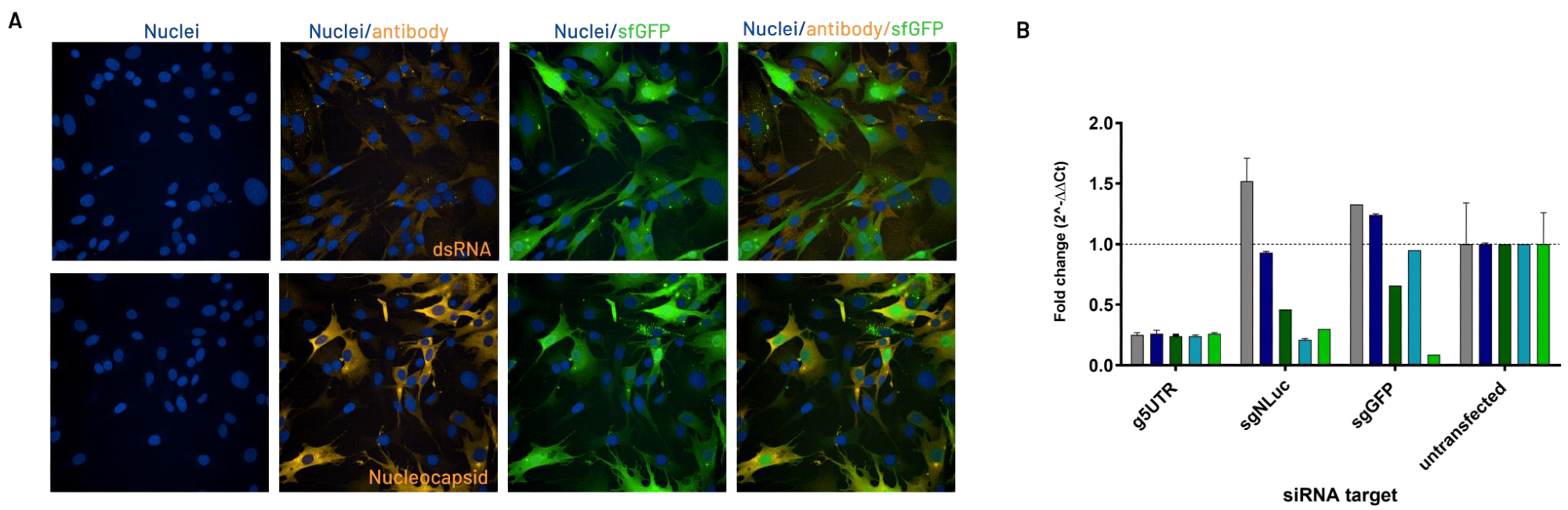
Fig. 3. The R484 Stable Cellular Model. Image Credit: Image courtesy of Cristina Alli et al., in partnership with ELRIG (UK) Ltd.
A stable cellular model was established for SARS-CoV replication using the SA062 replicon. This was achieved by transfecting BHK-21 cells with in vitro transcribed RNA and then applying antibiotic selection.
The resulting isolated cell clone, R484, was characterized using next-generation sequencing (NGS) on the total RNA extracted from the cells. This analysis confirmed the presence of the SA062 replicon and identified adaptive mutations that contribute to the replicon's persistence within the cells.
The presence of replicon RNA and the expression of viral proteins were demonstrated by detecting the presence of double-stranded RNA (dsRNA) and the Nucleocapsid protein in R484 cells (A). To validate the discontinuous transcription mechanism as a target in the cellular model, the team evaluated the effects of selectively knocking down genomic (g5'UTR) or subgenomic (sgNluc and sgGFP) RNAs using RT-qPCR (B).
Assay development and high-throughput screening campaign
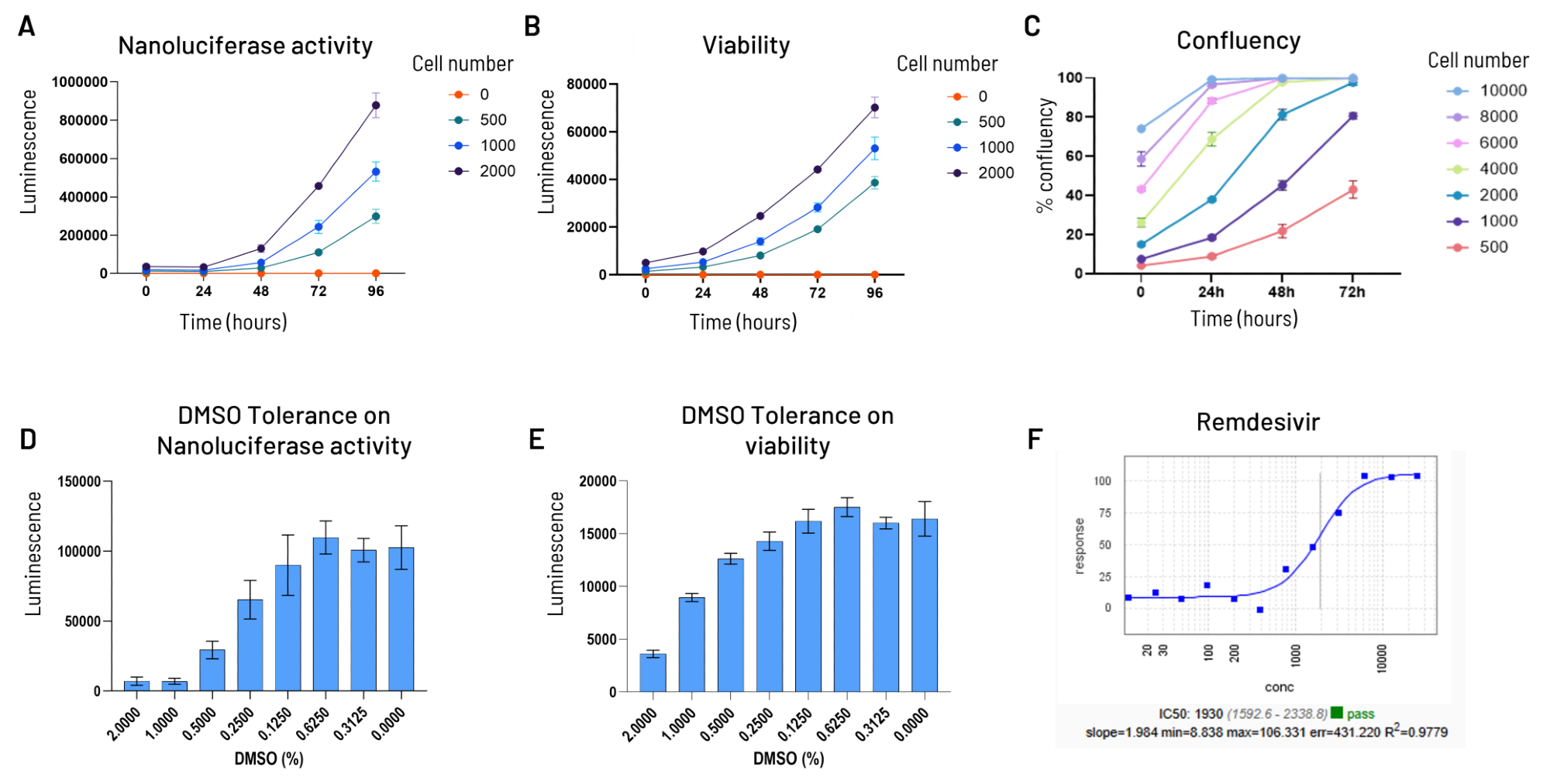
Fig. 4. Assay Set-up on R484 for HTS. Image Credit: Image courtesy of Cristina Alli et al., in partnership with ELRIG (UK) Ltd.
To be used in antiviral screening campaigns in a 384-well plate format, the R484 stable cellular clone was evaluated in terms of signal-to-background ratio of Nanoluciferase (A), cellular viability (B), density (C), DMSO sensitivity (D and E), and the Z-factor, and the treatment with a reference inhibitor (F).
The parameters optimized for the assay were 1500 cells/ well in the presence of 0.25 % DMSO and 72 hours of compound treatment. Applying these conditions, the Z-factor was greater than 0.5.
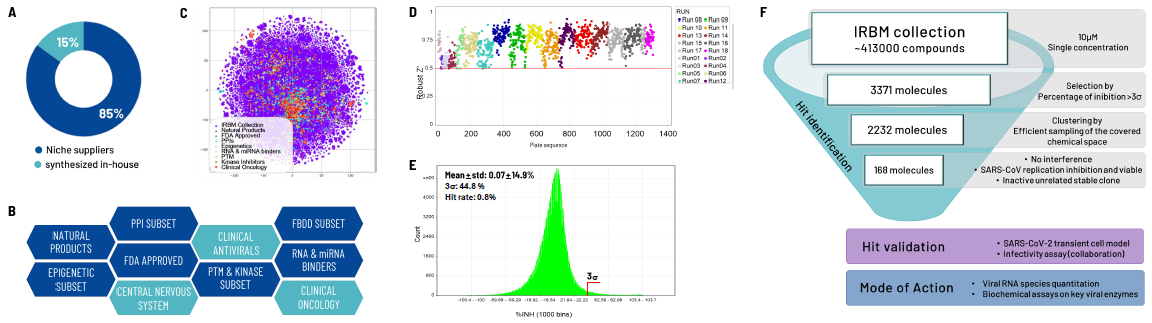
Fig. 5. Phenotypic HTS Campaign. Image Credit: Image courtesy of Cristina Alli et al., in partnership with ELRIG (UK) Ltd.
The IRBM compound collection comprises ~ 413,000 compounds, carefully curated from diverse sources, including niche and mainstream suppliers, as well as those synthesized in-house (A).
It is composed of a range of sub-libraries generated through in-house computational design and by acquiring compounds with demonstrated activity towards particular targets or areas of interest (B). The collection is balanced in terms of structural diversity and redundancy (C).
High-throughput screening was conducted by testing the entire IRBM collection at a single fixed dose concentration documented in R484. For all the tested plates, Robust Z' was > 0.5 (D).
Frequency distribution of % inhibition of the screened compounds was centered around zero, and using a cut-off corresponding to three seconds, 3371 hits were selected (E).
Following the identification of the most representative compounds through clustering, a total of 2232 molecules were chosen for dose-response curve testing. The potency (Efficacy Concentration 50 %, EC50) and cytotoxicity (Cytotoxic Concentration 50 %, CC50) were determined for 168 compounds.
Additionally, the team is currently conducting further characterization of their interference with the assay readout and their effects on an unrelated RNA virus (F).
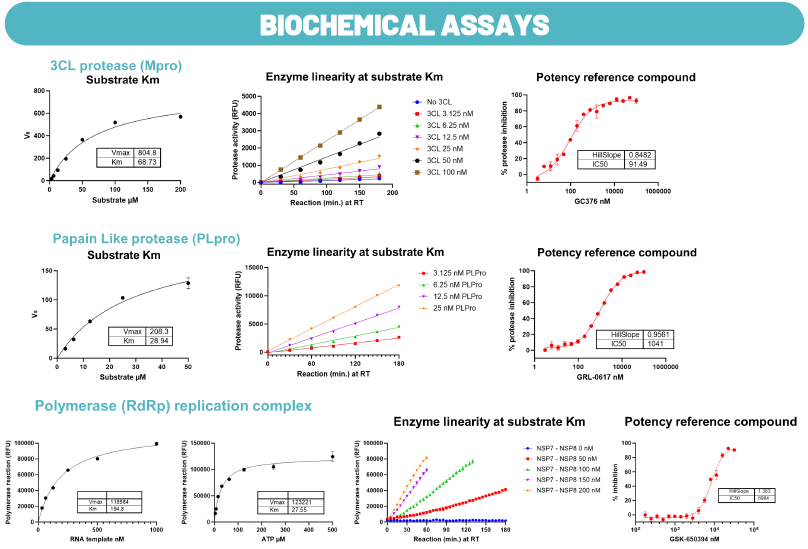
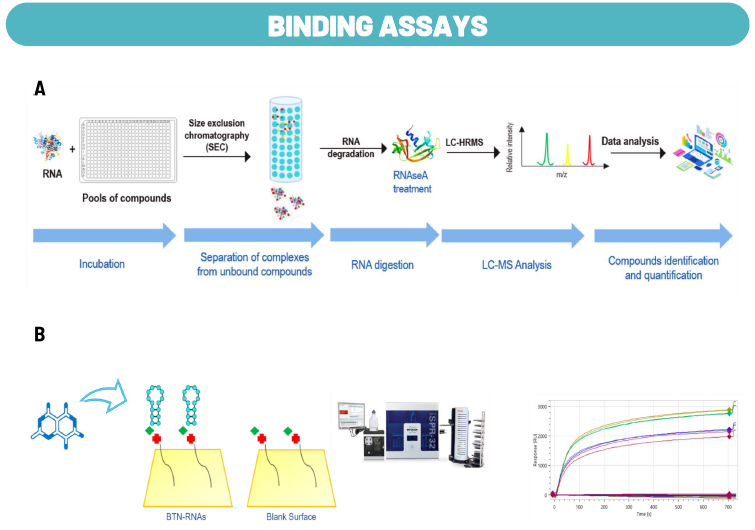
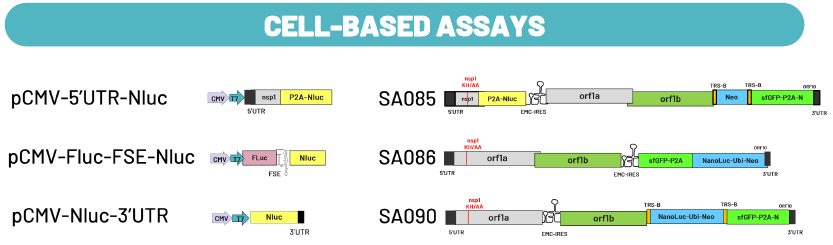
Fig. 6. Assays for Hit Characterization. Image Credit: Image courtesy of Cristina Alli et al., in partnership with ELRIG (UK) Ltd.
To characterize compounds from the HTS campaign, the research team has developed several assays with the aim of investigating their mechanism.
Biochemical assay: For the two viral proteases and the polymerase enzymes. For all the enzymes, they reported the Km calculation for the substrate, the enzymatic linearity at substrate Km, and the profile of the reference molecule.
Binding assay: Two in vitro RNA binding assays were developed, including an AS-MS approach (A), which allows for the identification of molecules that specifically interact with the viral 5'UTR in vitro, and the Surface Plasmon Resonance (SPR) as a refining technique to evaluate interactions with specific stem and loop structures (B).
Cell-based assay: To identify binders to RNA structures that are critical for viral replication, a panel of constructs was created. Specific viral regions, the 5'UTR and 3'UTR, were selected as involved in the discontinuous transcription process, along with unrelated RNA structures (i.e. the Frameshift Element, FSE), and were fused with a reporter gene to identify molecules that alter their functionality in cells specifically. The modified replicons were designed to discriminate for a specific mechanism of genome replication.
Conclusions
With this study:
- The team reported the first phenotypic stable cellular model of coronavirus replication.
- They demonstrated the possibility of identifying potent and selective compounds from the high-throughput screening campaign.
- An exhaustive panel of follow-up assays is currently being validated to enable an in-depth characterization of hits and the precise analysis of their mode of action.
About IRBM
IRBM is a leading contract research organization (CRO) based in Pomezia, near Rome, Italy.
The company specialises in integrated early-stage drug discovery across small molecules, peptides, and antibodies, providing end-to-end support from target validation and screening through to candidate nomination.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 13, 2025