This article and associated images are based on a poster originally authored by Divya Vemula and Vasundhra Bhandari and presented at ELRIG Drug Discovery 2025 in affiliation with the National Institute of Pharmaceutical Education and Research (NIPER).
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Background
Pseudomonas aeruginosa, a WHO-designated multidrug-resistant pathogen, causes severe hospital infections. With no vaccine against it, this study aims to design a multi-epitope vaccine to prevent infections caused by P. aeruginosa using reverse vaccinology and immunoinformatics.
The researchers have screened the full proteome (5,563 proteins) to identify non-human homologous, stable, non-allergenic, adhesive, and immunogenic proteins. Furthermore, shortlisted proteins (Probable coat protein A of bacteriophage Pf1, alkaline phosphatase L, and a secreted protein) were evaluated for B-cell, CTL, and HTL epitopes.
Six multi-epitope constructs (DV1–DV6) with different adjuvants were designed, followed by structural validation and assessment through docking with TLR2/TLR4 and molecular dynamics simulation studies. Among them, DV5 (HBHA-conserved adjuvant) showed the strongest receptor interactions, high stability, and 98.47 % predicted global HLA coverage. Hence, our results conclude that DV5 is a promising multi-epitope vaccine candidate against P. aeruginosa, utilizing reverse vaccinology in combating MDR pathogens.
Hypothesis
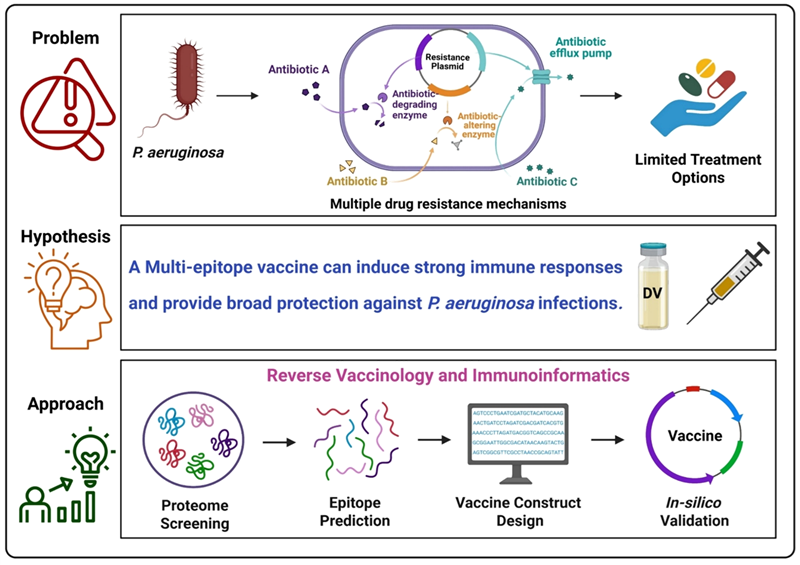
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
Methodology
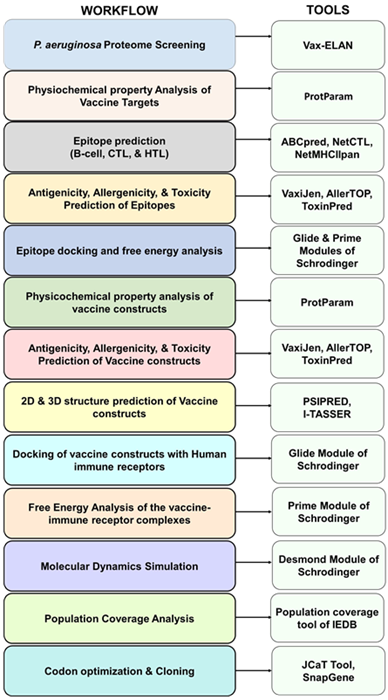
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
Results
A. P. aeruginosa proteome screening
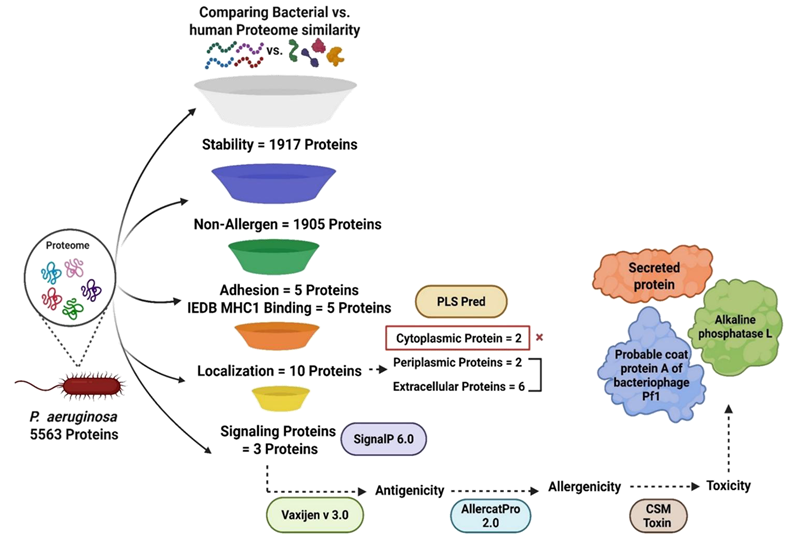
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
B. Physicochemical properties of vaccine targets
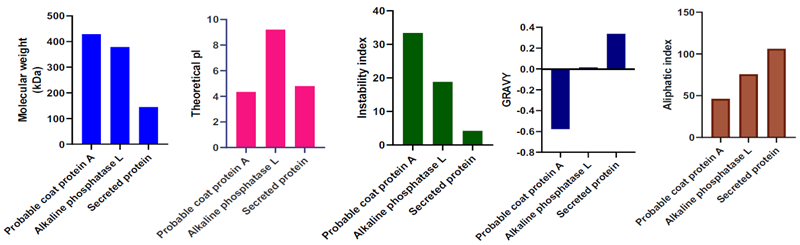
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
C. Epitope docking and free energy analysis
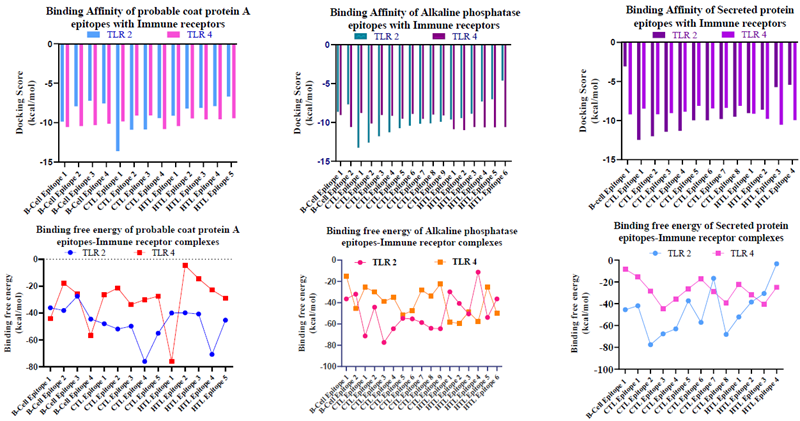
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
D. Multi-epitope vaccine construction
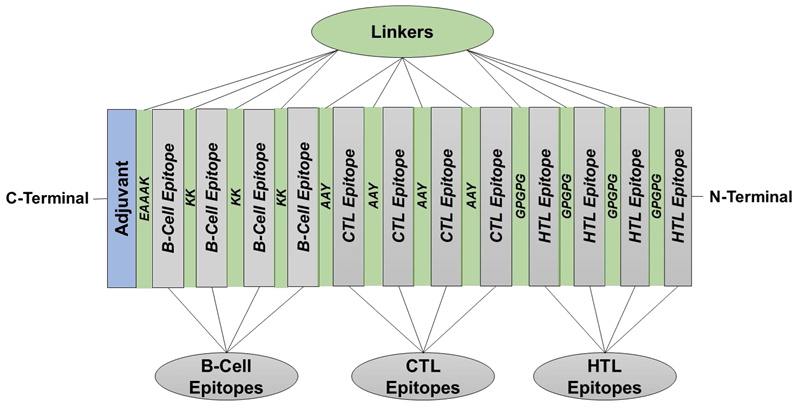
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
E. Physicochemical properties of vaccine constructs
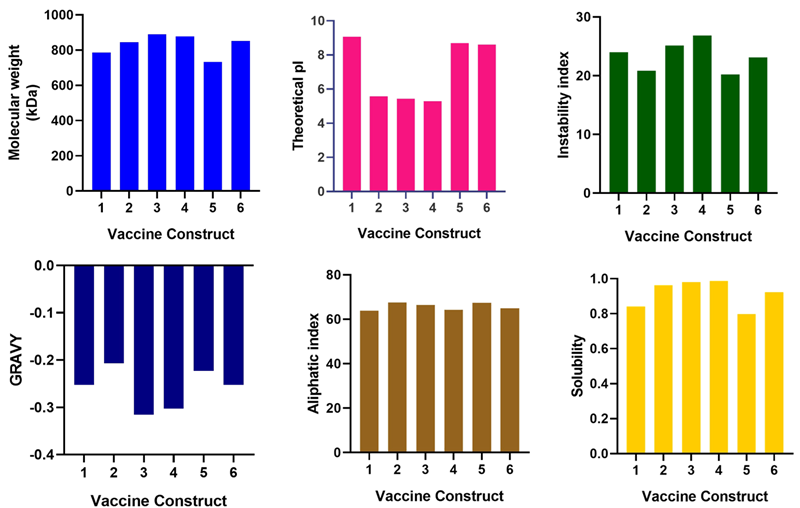
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
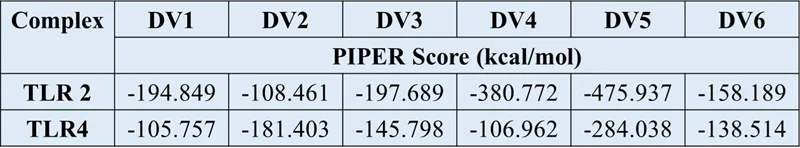
F. Docking of vaccine constructs with TLR2
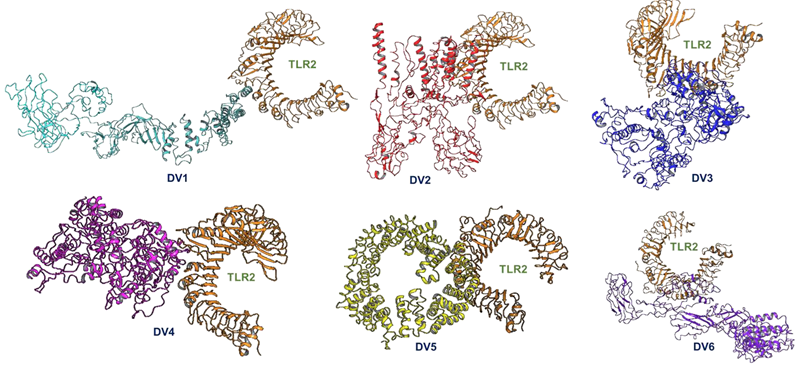
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
G. Docking of vaccine constructs with TLR4
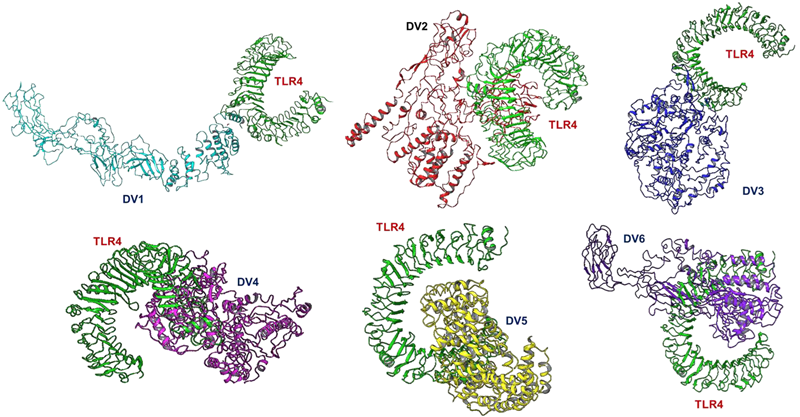
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
Source: ELRIG (UK) Ltd.
H. Molecular dynamics simulation of vaccine constructs with TLR2 & 4
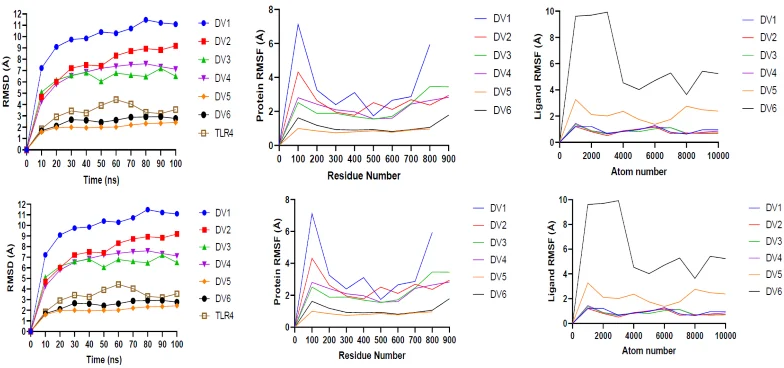
Image Credit: Image courtesy of Divya Vemula et al., in partnership with ELRIG (UK) Ltd.
RMSD and RMSF analysis confirmed DV5 as the most stable construct, with strong structural integrity and receptor binding.
I. Population coverage analysis
Source: ELRIG (UK) Ltd.
| Population |
MHC Class I & II combined |
| Coveragea |
Average hitb |
pc90c |
| World |
98.47 % |
2.82 |
1.72 |
a Projected population coverage
b Avg no. of epitope hits / HLA combinations recognized by population
c Min no. of epitope hits / HLA combinations recognized by 90 % of the population
K. Codon optimization & in-silico cloning
- DV5 (Designed Vaccine Construct 5; 2199 base pairs, Guanine–Cytosine content 60.48 %, Codon Adaptation Index 0.89) was codon-optimized using JCat for high-level expression in E. coli.
- The optimized gene was cloned into pET-28a(+) with an N-terminal 6xHis-tag under the T7 promoter. SnapGene validation confirmed the correct reading frame, restriction sites, and tag fusion, supporting efficient expression and purification. This construct bridges in silico vaccine design with experimental validation.
Conclusion
A multi-epitope vaccine against Pseudomonas aeruginosa was designed using reverse vaccinology and immunoinformatics. The construct included antigenic, nontoxic, non-allergenic epitopes with adjuvants to enhance immune response. It showed favorable physicochemical properties, stable immune receptor interactions, and broad HLA coverage.DV5 emerged as the most promising candidate for experimental validation.
References
- Priyamvada, P. and Ramaiah, S. (2023). Pan-genome and reverse vaccinology approaches to design multi-epitope vaccine against Epstein-Barr virus associated with colorectal cancer, Immunologic Research, 71(6), pp. 887–908. DOI: 10.1007/s12026-023-09403-2. https://link.springer.com/article/10.1007/s12026-023-09403-2.
- Ren, S. et al. (2019). Design and evaluation of a multi-epitope assembly peptide vaccine against Acinetobacter baumannii infection in mice, Swiss Medical Weekly, 149(2324), p. w20052. DOI: 10.4414/smw.2019.20052. https://smw.ch/index.php/smw/article/view/2627.
- Guedes, R.L.M. et al. (2018). A comparative in silico linear B-cell epitope prediction and characterization for South American and African Trypanosoma vivax strains, Genomics, 111(3), pp. 407–417. DOI: 10.1016/j.ygeno.2018.02.017. https://www.sciencedirect.com/science/article/pii/S0888754318301265?via%3Dihub.
- Li, J. et al. (2021). Reverse vaccinology approach for the identifications of potential vaccine candidates against Salmonella, International Journal of Medical Microbiology, 311(5), p. 151508. DOI: 10.1016/j.ijmm.2021.151508. https://www.sciencedirect.com/science/article/pii/S1438422121000370?via%3Dihub.
Acknowledgement
The author gratefully acknowledges the National Institute of Pharmaceutical Education and Research (NIPER), Hyderabad, and the Department of Pharmaceuticals, Government of India, for providing research infrastructure, computational facilities, and software support. The author also sincerely thanks the Department of Biotechnology (DBT), Government of India, under the Conference, Travel, Exhibition and Popular Lectures (CTEP) scheme, for travel grant support to present this work at ELRIG Drug Discovery 2025, Liverpool, UK.
About NIPER Hyderabad
The National Institute of Pharmaceutical Education and Research (NIPER), Hyderabad, is an Institute of National Importance, established with the objective of becoming a centre of excellence for advanced research in pharmaceutical sciences.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Jan 6, 2026