This article and associated images are based on a poster originally authored by Matthew Chung, William Guesdon, Kai Lawson-McDowall and Matthew Alderdice and presented at ELRIG Drug Discovery 2025 in affiliation with Sonrai Analytics.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Sonrai Biodiscovery is a platform designed to advance tertiary and functional analysis for hypothesis generation and testing. Despite the richness of modern data, transforming complex multimodal datasets into clear, contextualized insight remains one of the key challenges in drug discovery.
While omics, imaging, and functional assays are now routine, biomedical data analysis has struggled to keep pace with the rapid generation of biological knowledge and innovative analytical methods. Current tertiary analysis approaches remain fragmented and slow, often relying on generic pathway or ontology annotations that fail to reflect biological context. This gap limits translational programs, as valuable datasets lose impact due to poor interpretability and disconnected prior knowledge.
Sonrai Biodiscovery directly addresses this by enabling context-enriched multimodal analytics that integrate multiple results, leverage prior knowledge, and connect securely within a compliant cloud environment. It enhances scientists’ ability to generate well-informed hypotheses while laying the foundation for agentic, knowledge-driven discovery workflows.
Who is Sonrai?
Their mission is to power the most important precision medicine breakthroughs through connected data and collaborative science.
Sonrai helps teams to maximize their data by
- Enabling collaborative tools for data visualization and analysis
- Integrating data silos into a permanent, compliant infrastructure
- Ensuring auditability, reproducibility, and traceability
- Reducing cost, time, and risk from early discovery to clinical trials.
1. Knowledge-enriched
Prior knowledge should play a central role in tertiary and functional analysis. The scientific community has already captured vast biological understanding in data form, including relational databases and molecular interaction networks. Single-cell RNA-seq atlases provide sources of signature matrices for deconvolution.
Contextual ligand-receptor networks, interactomes, and gene regulatory maps facilitate the inference of mechanisms. Drug-disease relationships in medical knowledge graphs, along with perturbation data, reveal clinical relevance in underlying biology. This approach, now popularized through foundation models, can also be applied methodically beyond machine learning prediction tasks to generate testable hypotheses.1
The team's upstream analysis performs ligand activity inference by aligning ligand target predictions with observed expression changes, adapted from NicheNet, and enhanced using Accurate Precision ranking. They further integrate Maximum Flow analysis on receiver cell-specific hybrid interactome-gene regulatory networks to infer transcription factor activity, comparable to interFLOW.2,3
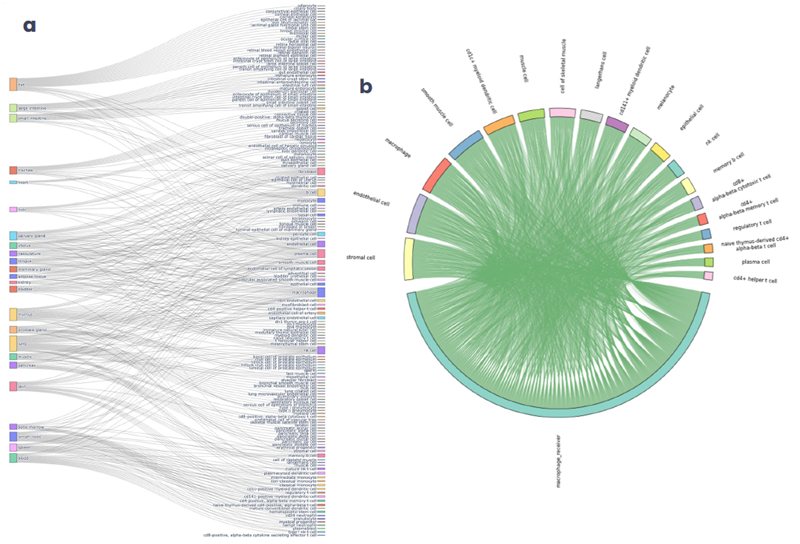
Figure 1. Prior knowledge networks and ontologies present a wealth of contextual information. (a) Sankey plot showing the connected tissue and cell type ontologies of our curated interactomes. (b) Chord diagram showing the complete set of ligand-receptor interactions from skin cell types to macrophages. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
2. Contextual analytics
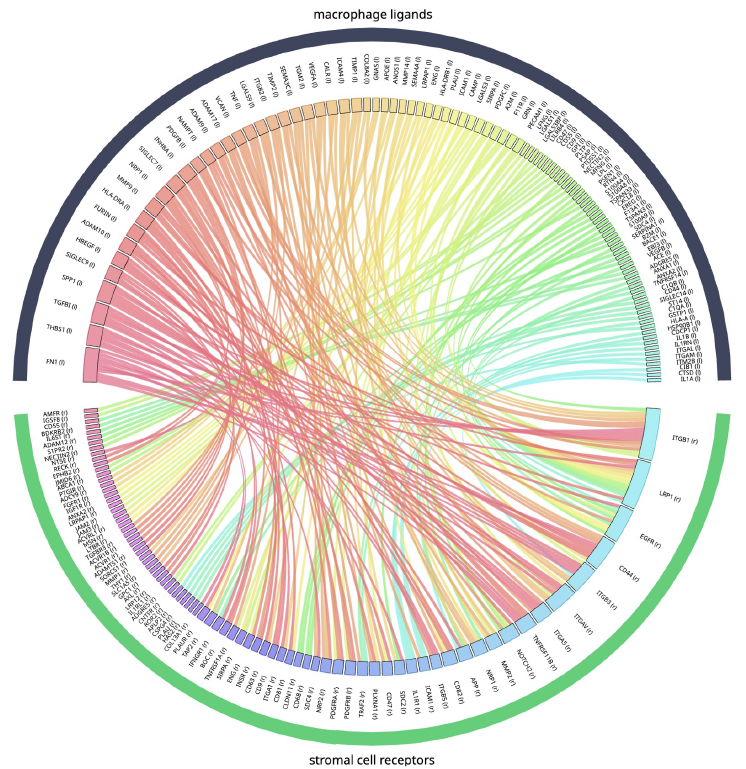
Figure 2. Chord diagram showing the results of ligand-receptor interaction, restricted to only include proteins from macrophages and stromal cells, and filtered for their presence in the data. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
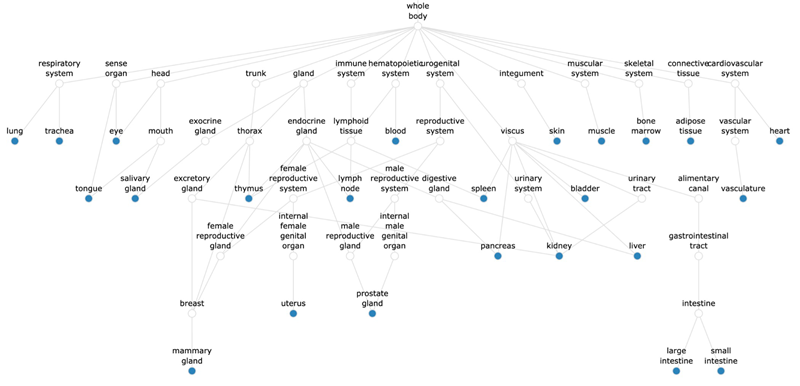
Figure 3. Flow diagram showing the hierarchical relationships of tissues from which we analyzed their cell types from large-scale atlases. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
The same proteins can play distinct roles across tissues and cell types. Recognizing this, the team incorporated as much biological context as possible into each analysis. Enabled by single-cell omics and large-scale atlases, the researchers constructed cell type-specific interactomes, inspired by PINNACLE (Figure 3).4,5,6 These interactomes represent normal molecular activities unique to each cell type. This contextual information enriches upstream analysis and reduces noise (Figure 2).
They also facilitated cell-type deconvolution of bulk data by providing validated tissue-specific signature matrices from atlas-scale sources, implemented via a Python-based ν-SVR algorithm.7,8 Cell-type proportions derived in this way offer a powerful contextual lens for interpreting gene expression changes.
3. Multimodal by design
Modern biology should be studied across modalities that capture complementary layers of information. Drug discovery projects often produce multi-omic, imaging, and metabolomic data that are rarely analysed together. Sonrai Biodiscovery, built on the multimodal data analysis framework MuData9 (Figure 7), enables the flexible integration of diverse data types, as demonstrated in single-cell and spatial multiomics.10,11
The team recommends using MOFA12 to extract latent factors across modalities, extending it within Biodiscovery to include diagnostics that link factor composition with clinical metadata (Figure 4).
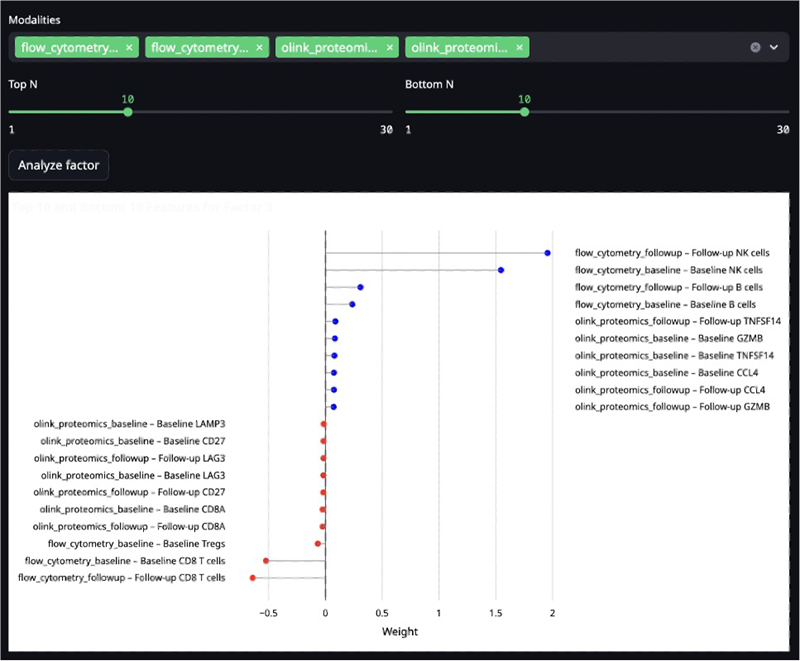
Figure 4. Screenshot of the interactive MOFA. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
Metabolism, often siloed in analysis, is also integrated. Gene expression results are mapped to enzymatic reactions in human metabolism, which can be further augmented when metabolomics data is available (Figure 5). This interconnected representation also supports advanced algorithms, such as GNN-based metabolic flux prediction from expression data.13
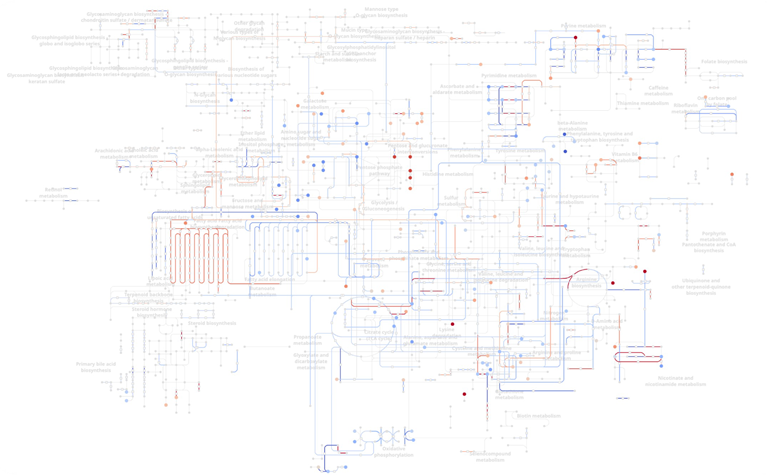
Figure 5. Human metabolism network overlaid with differential gene expression and metabolite abundance. Each node (dot) is a metabolite, while each edge (line) is gene(s) that perform the enzymatic reaction. Red indicates upregulation in macrophages of skin cancer immunotherapy responders, while blue indicates downregulation. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.

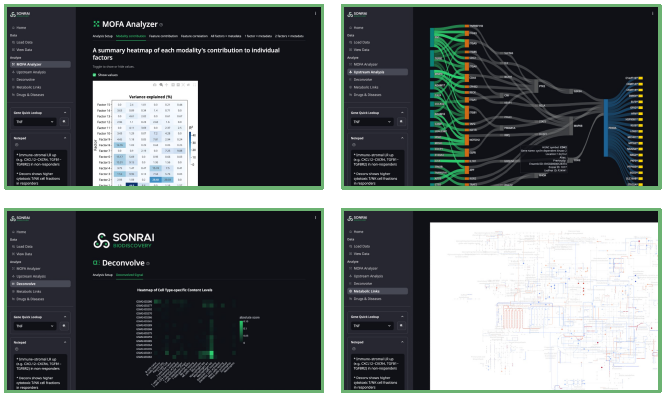
Figure 6. Screenshots of the Sonrai Biodiscovery user interface. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
4. Cross-analytic integration
Combining outputs from diverse analytical tools can reveal higher-order biological insights not visible from individual analyses alone. Yet, this has historically been time-consuming and technically fragmented. Sonrai Biodiscovery resolves this by adopting the MuData framework (Figure 7), which enables seamless storage and cross-analysis of multiple results.9
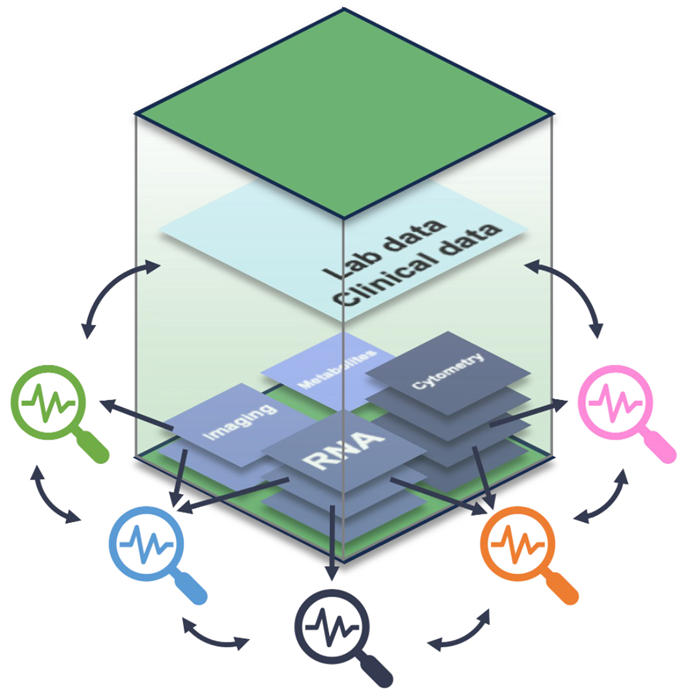
Figure 7. Schematic diagram of cross-analytic flexibility enabled by the MuData multimodal framework. Specialized analyses can be performed on one or more data modalities, but their output can also be analyzed with other outputs and with clinical and experimental observations. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
Scientists can statistically compare and visualize integrated results (Figure 8), for example, linking deconvolution-derived cell-type proportions or MOFA latent factors to clinical outcomes. This empowers scientists to ask deeper, context-driven questions that drive hypothesis generation and mechanistic understanding.
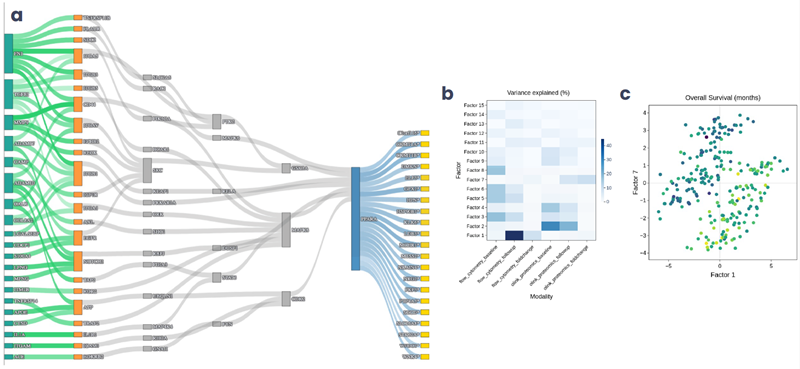
Figure 8. (a) Upstream plot created through the integration of ligand-receptor and transcription factor activity analysis, based on prior knowledge networks, and filtered for cell type context. (b) Heatmap from multi-omics factor analysis showing the contribution of each data modality to the final latent factors. (c) Scatterplot from the same analysis showing how two multi-omic factors segregated patients by overall survival in a cancer clinical study. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
5. Securely connected
Sonrai Biodiscovery is embedded within the broader Sonrai ecosystem, connecting seamlessly with compliant, production-ready infrastructure. Sonrai Pipelines delivers automated and auditable omics data processing. Results such as count matrices and differential gene expressions can be accessed in Biodiscovery. Sonrai Imaging supports pathology-optimized AI/ML workflows using foundation models.
Whole-slide and tile embeddings generated can be used in Biodiscovery for multimodal analysis. For advanced AI/ML, Sonrai Studio offers cloud-native environments for model development, complementing tertiary analyses. Sonrai Insights centralizes visual dashboards for analytics and collaborative reporting.
All modules operate within each client’s private Sonrai Cloud, equipped with scalable compute, granular access control, private workspaces, project management, and a central data repository managed under ISO-certified quality and cybersecurity standards.
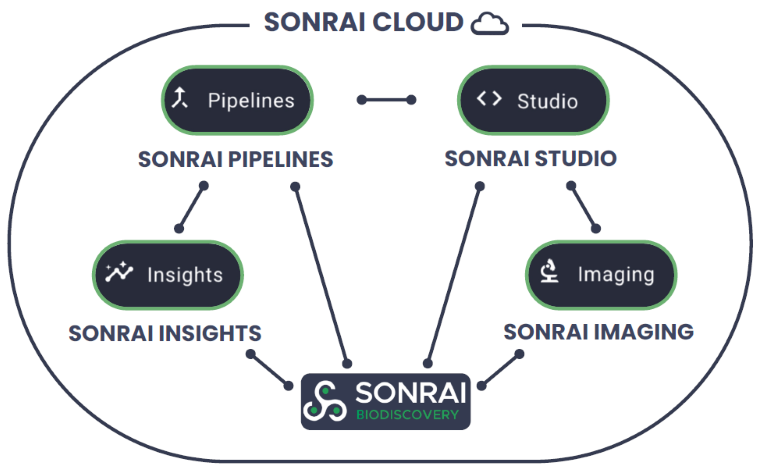
Figure 9. Schematic diagram of Sonrai Biodiscovery interconnecting with other Sonrai platform features within a secure cloud environment. Image Credit: Image courtesy of Matthew Chung et al., in partnership with ELRIG (UK) Ltd.
Continuous development
Sonrai Biodiscovery is built to evolve with emerging methodologies in tertiary analytics. Its modular, multimodal framework readily accommodates new data types and integrates cutting-edge algorithms. The company is actively seeking scientists’ feedback on how to better empower them with context-aware, knowledge-enriched, and intuitive analytics.
References
- Chandak, P., Huang, K. and Zitnik, M. (2023). Building a knowledge graph to enable precision medicine. Scientific Data, (online) 10(1), p.67. https://doi.org/10.1038/s41597-023-01960-3.
- Chananchida Sang-aram, Browaeys, R., Seurinck, R. and Yvan Saeys (2025). Unraveling cell–cell communication with NicheNet by inferring active ligands from transcriptomics data. Nature Protocols. https://doi.org/10.1038/s41596-024-01121-9.
- Sheinin, R., et al. (2024). interFLOW: maximum flow framework for the identification of factors mediating the signaling convergence of multiple receptors. npj Systems Biology and Applications, 10(1). https://doi.org/10.1038/s41540-024-00391-z.
- Abdulla, S. et al. (2024). CZ CELLxGENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data, Nucleic Acids Research, 53(D1), pp. D886–D900. https://doi.org/10.1093/nar/gkae1142.
- Rood, J.E., et al. (2024). The Human Cell Atlas from a cell census to a unified foundation model. Nature. (online) https://doi.org/10.1038/s41586-024-08338-4.
- Li, M.M., et al. (2024). Contextual AI models for single-cell protein biology. Nature Methods. (online) https://doi.org/10.1038/s41592-024-02341-3.
- Newman, A.M., et al. (2019). Determining cell type abundance and expression from bulk tissues with digital cytometry. Nature Biotechnology, 37(7), pp.773–782. https://doi.org/10.1038/s41587-019-0114-2.
- Nava, A., et al. (2023). Novel evaluation approach for molecular signature-based deconvolution methods. Journal of Biomedical Informatics, 142, p.104387. https://doi.org/10.1016/j.jbi.2023.104387.
- Bredikhin, D., Kats, I. and Stegle, O. (2022). MUON: multimodal omics analysis framework. Genome Biology, 23(1). https://doi.org/10.1186/s13059-021-02577-8.
- Wolf, F.A., Angerer, P. and Theis, F.J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biology, 19(1). https://doi.org/10.1186/s13059-017-1382-0.
- Palla, G., et al. (2022). Squidpy: a scalable framework for spatial omics analysis. Nature Methods, 19(2), pp.171–178. https://doi.org/10.1038/s41592-021-01358-2.
- Argelaguet, R., et al. (2018). Multi‐Omics Factor Analysis - a framework for unsupervised integration of multi‐omics data sets. Molecular Systems Biology, (online) 14(6). https://doi.org/10.15252/msb.20178124.
- Alghamdi, N., et al. (2021). A graph neural network model to estimate cell-wise metabolic flux using single-cell RNA-seq data. Genome Research, (online) 31(10), pp.1867–1884. https://doi.org/10.1101/gr.271205.120.
About Sonrai Analytics
Sonrai Analytics was established to meet a critical need in precision medicine, addressing the challenge of integrating large-scale multi-omic data to uncover meaningful insights and hypotheses that drive the discovery of new therapies and biomarkers. Guided by a commitment to improving diagnosis and treatment for patients worldwide, Sonrai’s purpose is to turn complex data into actionable breakthroughs.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 13, 2025