international HUPO, Bruker Corporation today announced new machine learning additions to Bruker ProteoScape™ software for immunopeptidomics and PTM analysis. In addition, new OmniScape™ workflows enable rapid and confident de novo identification and proteoform characterization. Augmenting the launch of the timsOmni™, the integration of IPT’s SampleStream™ technology delivers high-throughput top-down proteoform analysis, combining sample recovery and cleanup for advanced biopharma research.
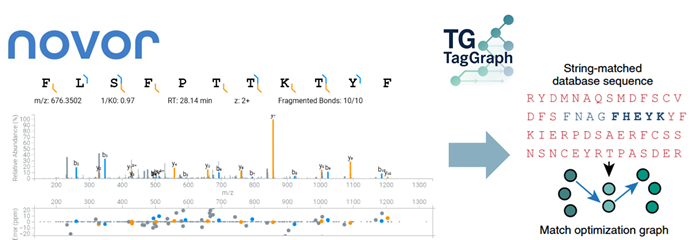 Figure 1. Machine learning-based de novo sequencing in conjunction with mapping of PTMs for confident peptide variant identification. Image Credit: Bruker
Figure 1. Machine learning-based de novo sequencing in conjunction with mapping of PTMs for confident peptide variant identification. Image Credit: Bruker
A. ProteoScape™ integrates advanced machine learning with TIMS DIA-NN, Spectronaut® 20, for immunopeptidomics, PTM, and de novo neoepitope discovery.
Bruker’s ProteoScape™ platform now features several additions focused on immunopeptidomics and posttranslational modifications (PTMs), providing high-throughput and high-sensitivity proteomics, with Spectronaut® 20 (SN20). SN20 offers the new Kuiper search engine, specifically designed for immunopeptidomics and PTM searches.
Prof. Dr. Oliver Schilling at the Institute for Surgical Pathology, Medical Center - University of Freiburg, Freiburg, Germany, said, ”The Bruker ProteoScape™ platform, with integrated Spectronaut and TIMS DIA-NN, has significantly boosted our ability to reliably analyze patient-derived samples for precision oncology. We can now perform precise and comprehensive analyses of complex clinical samples in real time, enabling us to rapidly turn mass spectrometry data into a powerful resource with the potential to directly impact medical decisions. Combined with the ProteoScape Novor module, this setup makes it easy to handle and confidently analyze de novo peptides, which has been a game-changer for our immunopeptidomics research and a significant step towards deeper understanding of the immune landscape in cancer."
In addition, ProteoScape™ now features TagGraph from the Elias Lab at the CZ Biohub, which provides scalable neopeptide discovery by aligning de novo sequencing results from
Trade news
ProteoScape™ Novor with sequence databases, introducing modifications and amino acid substitutions to uncover modified peptides. Together with novel visualization of peptide motifs these tools enable accurate analysis of HLA-presented peptides from lowest sample input.
Dr. Joshua Elias, CZ Biohub: "By integrating TagGraph into Bruker ProteoScape on the timsTOF platform, we’ve helped automate open modification searches in mass spectrometry-based proteomics. This implementation streamlines de novo sequencing alignment, significantly expanding peptide identifications and introducing modifications and amino acid substitutions as needed. Benchmarking against standard DB tools demonstrated a 30 % increase in assigned spectra, with at least 20 % of novel peptides carrying unexpected modifications. This advancement makes open modification searches more accessible, helping establish them as a standard feature in proteomics workflows."
Trade news
B. Novel glycoproteomics de novo workflows with timsOmni™ and OmniScape enable confident proteoform characterization
With OmniScape version 2026, Bruker introduces significant improvements for its proteomics de novo workflow. Notable advancements encompass the residue-specific score and visualization via a confidence sequence heatmap for results validation. In addition, the unique advantage of electron-based activation (ECD, EXD, and EID) on the timsOmni™ has been demonstrated for comprehensive glycopeptide analysis. Extended glycan structures were preserved on the peptide backbone, allowing further investigation into biologically highly relevant glycopeptide topology through MSn experiments (Figure 2). The versatility of timsOmni™ enables scientists to precisely localize glycan moieties and gain detailed insights into PTMs, advancing both structural and functional understanding of biological networks.
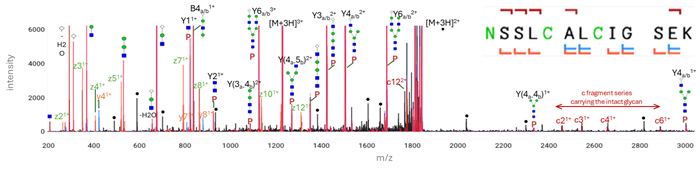
Figure 2. ECciD on the timsOmni™ – transferrin peptide analysis revealing glycan structures and preserved peptide backbone information. Image Credit: Bruker
Dr. Francisco Alberto Fernandez Lima, FIU, USA: “Our laboratory has extensively utilized OmniScape for de novo and Top-Down protein-PTM sequence validation from ExD and UVPD datasets from timsTOF and MRM-MS instruments. Different from alternative approaches, OmniScape is more user friendly and allows for interactive fragment assignment/confirmation of proteoforms including variable AA level PTMs, m/z internal calibration and MS-BLAST search for database inquiries.”
C. Bruker and IPT launch SampleStream™–timsOmni™ integration for automated, high-throughput intact proteoform profiling.
The SampleStream™ technology from IPT enables and streamlines the analysis of proteoforms by mass spectrometry, delivering exceptional sample recovery and efficient cleanup. The combination of MSn enrichment on the timsOmni™ with the SampleStream™ technology is uniquely suited for high-throughput proteoform analysis.
Trade news

Figure 3. IPT SampleStream™ integrated into a Bruker Elute HT coupled to the timsOmni™. Image Credit: Bruker
Together, we see significant opportunity to bring high-throughput, intact and top-down protein analysis to a broader global market. Integrating SampleStream with Bruker’s recently released timsOmni system represents a powerful combination that could accelerate innovation and adoption of these data types across proteomics, biopharma, and clinical research.”
Dr. Philip Compton, CEO, Integrated Protein Technologies, Inc.
Daniel Hornburg, Vice President, Biomarkers & Precision Medicine, Bruker. “Proteomics is evolving rapidly beyond canonical protein maps. With Bruker’s innovations, we’re pushing the boundaries of bottom-up analysis for example for PTMs and cancer neoantigens, and at the same time, making proteoform profiling more efficient, accessible and confident. By closing the gap between these worlds, we’re giving scientists the tools to see biology as it truly operates and study health and disease.”