Functional protein microarrays serve as potent tools for immunoprofiling, offering valuable insights into the disease state through the distinct repertoire of antibodies present in individuals. However, acquiring this information necessitates a well-designed protein microarray.
Functional protein microarrays emerged in the early 2000s, lagging slightly behind DNA microarrays. Both technologies were developed to facilitate high-throughput analysis but employed different detection techniques.
DNA microarrays employ miniaturized hybridization assays, while functional protein microarrays utilize miniaturized indirect immunofluorescent assays. In the case of functional protein microarrays, proteins are printed onto a planar surface.
Antibodies, which carry an individual's disease history, circulate in the blood and lymph.The test sample for functional protein microarrays is typically serum replete with host antibodies that bind to complementary proteins. To visualize the protein-serum antibody complex, a fluorescently tagged secondary antibody, specific to the serum antibody species, is applied.
Functional protein microarrays can encompass numerous proteins, fragments, or peptides, enabling immunoprofiling of individuals and populations, thereby identifying distinctive immune experiences, including early-stage diseases. Functional protein microarrays heavily rely on the specificity of antigen-antibody binding.
In biological systems, proteins necessitate a precise three-dimensional configuration to carry out their functions. During translation, the ribosome synthesizes a linear chain of amino acids, which subsequently folds into a secondary structure due to hydrogen bonding between neighboring amino acids.
The protein further folds into its tertiary structure, driven by various forces such as hydrogen bonds, van der Waals forces, and hydrophobic interactions. This folding process brings distant amino acids and different atomic groups that constitute the peptides into proximity.
Each atomic group represents a distinct section of the protein, and upon folding, these groups align to form the epitope—a unique surface recognized by a specific antibody (see Figure 1).
Due to the non-continuous nature of amino acids in the tertiary structure, the resulting antibody binding site is referred to as a discontinuous epitope (Figure 2) (Barlow et al., 1986; Van Regenmortel, 1996).
The epitope consists of atom groups rather than a linear amino acid sequence. To visualize the epitope and illustrate its discontinuity, X-Ray crystallography has been employed.
If the protein undergoes unfolding, denaturation, or fragmentation, the epitope is lost (Van Regenmortel, 2006, 2016). Several factors, including temperature, pH, other molecules, and the underlying amino acid sequence, can affect protein folding.
Intracellular chaperones and folding enzymes play a crucial role in ensuring the proper folding of the protein into its functional three-dimensional structure. In certain cases, proteins consist of subunits that must assemble for functionality.
Ninety percent of antibodies recognize the discontinuous epitopes formed through protein folding (Van Regenmortel, 1996).
This poses a challenge in manufacturing protein microarrays since retaining these epitopes during production and immobilization of correctly folded proteins is difficult to control in a manufacturing environment.
Epitope loss results in the disappearance of the antibody recognition site, leading to increased non-specific binding and higher rates of false positives and false negatives. Sengenics KREX protein folding technology is specifically designed to ensure that the protein array comprises properly folded, full-length proteins.
In this patented technology, a biotin carboxyl carrier protein (BCCP) is fused in-frame with each array protein as a folding marker. Misfolded or fragmented proteins cause misfolding of BCCP, which masks its biotinylation site and prevents it from binding to the streptavidin-coated array surface (Figure 3). This technology preserves discontinuous epitopes and facilitates optimal antibody-epitope binding.
Techniques like protein microarrays utilize the well-established method of indirect immunofluorescence to decode this history. By employing technology that addresses the nature of antibody-antigen binding, reliable results with a high signal-to-noise ratio are ensured.
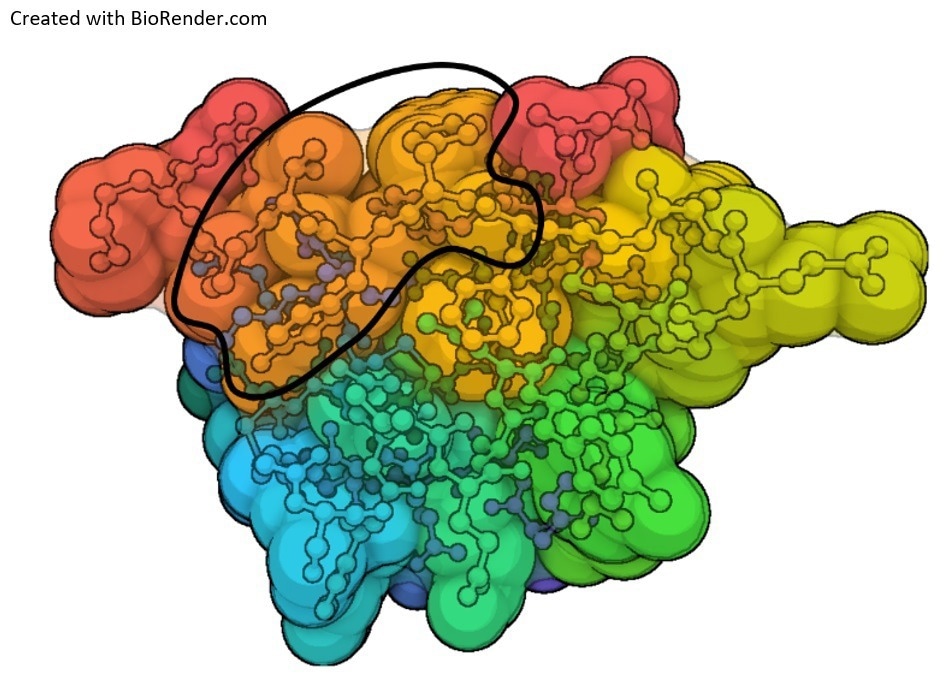
Figure 1. Antibody epitopes form from different atomic groups from discontinuous amino acids that form a surface (outline) when the protein folds. A contact map of insulin is shown below with atomic groups overlayed. Blue, orange, and dark orange atomic groups fold over each other to form the epitope. Image Credit: Sengenics
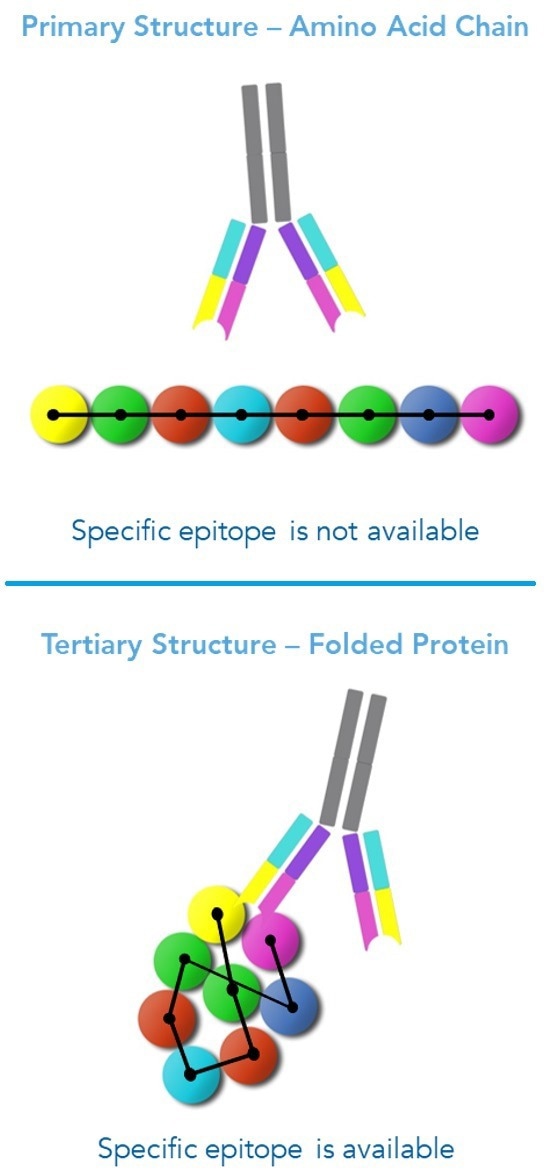
Figure 2. Antibodies recognize discontinuous epitopes formed during protein folding. When a protein folds, non-contiguous amino acids come within close proximity, forming a unique epitope binding site for antibody binding. Image Credit: Sengenics
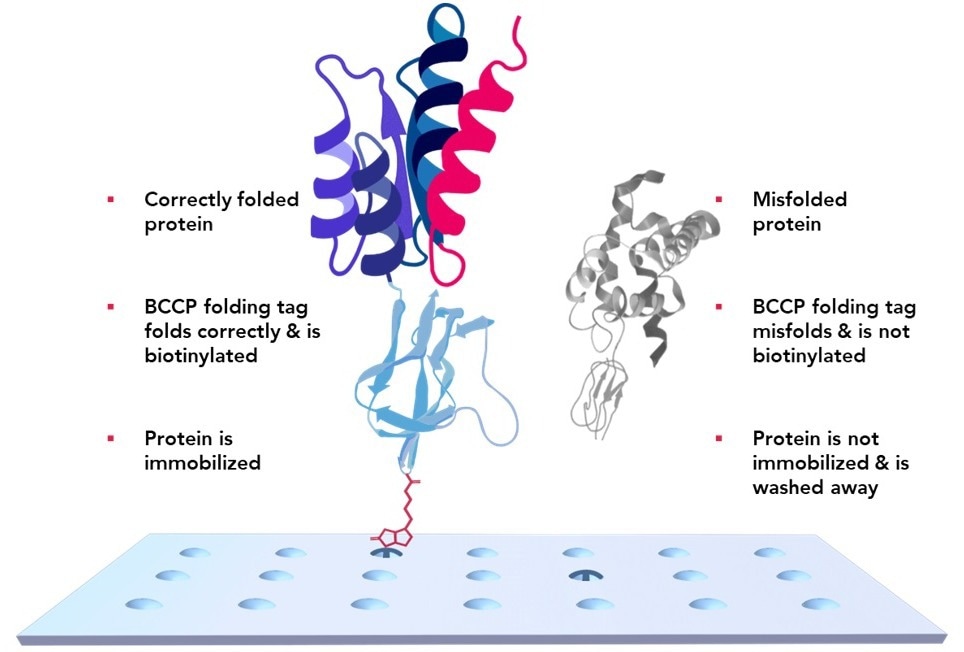
Figure 3. KREX Technology ensures only full-length properly folded proteins adhere to the array surface. A Biotinylated Carboxyl Carrier Protein is coded in-frame with the desired protein. The slide surface is printed with a hydrogel containing streptavidin. If a protein misfolds, the BCCP misfolds with it and cannot bind to the streptavidin array surface. Misfolded proteins are washed away. Image Credit: Sengenics
References and further reading
- Barlow, D. J., Edwards, M. S., & Thornton, J. M. (1986). Continuous and discontinuous protein antigenic determinants. Nature, 322(6081), 747-748. https://doi.org/10.1038/322747a0
- Van Regenmortel, M. H. (2006). Immunoinformatics may lead to a reappraisal of the nature of B cell epitopes and of the feasibility of synthetic peptide vaccines. J Mol Recognit, 19(3), 183-187. https://doi.org/10.1002/jmr.768
- Van Regenmortel, M. H. (2016). Structure-Based Reverse Vaccinology Failed in the Case of HIV Because it Disregarded Accepted Immunological Theory. Int J Mol Sci, 17(9). https://doi.org/10.3390/ijms17091591
- Van Regenmortel, M. H. V. (1996). Mapping Epitope Structure and Activity: From One-Dimensional Prediction to Four-Dimensional Description of Antigenic Specificity. Methods, 9(3), 465-472. https://doi.org/10.1006/meth.1996.0054
About Sengenics
Sengenics is an immunoproteomics company working to improve patient outcomes through physiologically relevant, data-guided decision making. Our solutions enable the discovery and validation of autoantibody biomarker signatures for patient stratification, therapeutic response prediction and elucidation of disease mechanisms.
The company has a global footprint with multiple corporate and research sites across the world with customers and collaborators that include top pharma, biotech and ivy league academic institutions in North America, Europe, and Asia.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.