One method for visualizing dynamic biological processes is single particle tracking, a localization microscopy method. For this technique to be successful, several factors need to be considered, including laser power, fluorophores, and experimental design.
This article outlines a range of labeling techniques, including genetically encoded labels, non-genetically encoded labels, and live RNA tracking, as well as their benefits for different experimental requirements.
General considerations for single particle tracking
Single particle tracking involves the detection and localization of spots over time. Localization accuracy depends on the square root of the number of photons collected per spot: brighter spots can be localized with higher accuracy, enhancing the quality of single particle tracks.
Brightness is even more important for single particle tracking than other localization methods, as both spatial and temporal resolution are very important.
For fast-moving particles or rapidly evolving processes, very high frame rates are sometimes required to prevent blurring, resulting in shorter photon collection windows. In these conditions, spot brightness is even more crucial, and increased excitation intensity can raise the number of photons collected per spot.
The Bruker Vutara VXL super-resolution microscope is equipped with watt-scale lasers for most imaging lines, providing highly intense, uniform illumination across the whole field of view. However, high-intensity illumination can cause rapid spot bleaching, so a balance is required between intensity and duration for single particle tracking.
With Vutara VXL’s various neutral density filters, stable illumination levels, ranging from milliwatts to kilowatts per square centimeter, can be achieved on the same system with the same laser line, and even within a single experiment if desired.
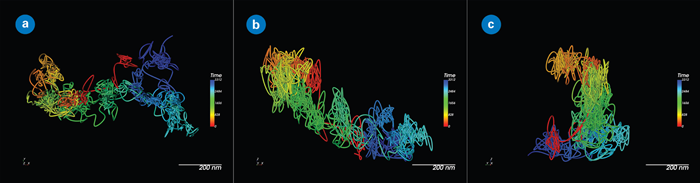
Figure 1. A single QD655-labeled membrane protein extracellular domain, showing the importance of 3D tracking for accuracy, even in relatively flat contexts: (a) XY view; (b) XZ view; and (c) YZ view. Image Credit: Nano Surfaces and Metrology
General guidelines for labeling strategy selection
In most cases, fluorophores with strong photostability are favored for labelling, meaning they should resist bleaching, blinking, or photoconversion.
This contrasts with other single-molecule localization microscopy (SMLM) methods, where blinking and photoconversion are advantageous. In most cases, quantum dots represent the brightest and most photostable labels available.
However, it can be difficult to attach these labels to the correct structure or material, as they must be introduced into the cell already connected to the protein or object of interest, linked to an antibody, or directly ligated via click chemistry.
Quantum dots are often too large to pass freely through the membrane, particularly when attached to antibodies. When targeted against intracellular components, this requires introducing the cell through microinjection, transfection, or other disruptive approaches.
Another factor is ease of use. Genetic encoding offers a convenient alternative in certain situations, streamlining labeling since plasmid transfection is usually less complex than creating and delivering quantum dot-labeled proteins.
However, such genetic labels have limitations. For example, they require sparse labeling (only a few particles per cell) to enable tracking. One way to achieve sparse labeling is endogenous tagging of sparse structures (e.g., centrosome or specific mRNAs) or low-expressing, low-copy-number plasmids.
SPT-PALM (single-particle tracking photoactivated localization microscopy) offers an additional sparse labeling option by photoconversion or photoactivation of a subset of dyes by brief pulses of photoactivation, followed by longer tracking and recording.
Choosing the most suitable approach and label for experiments can be challenging. The Bruker applications team can provide tailored guidance on specific experimental questions.
Bruker’s Vutara VXL tools are compatible with all techniques described, and its robust SRX software can image and monitor particles, whether they are quantum dots, fluorescent proteins, or single dye molecules.
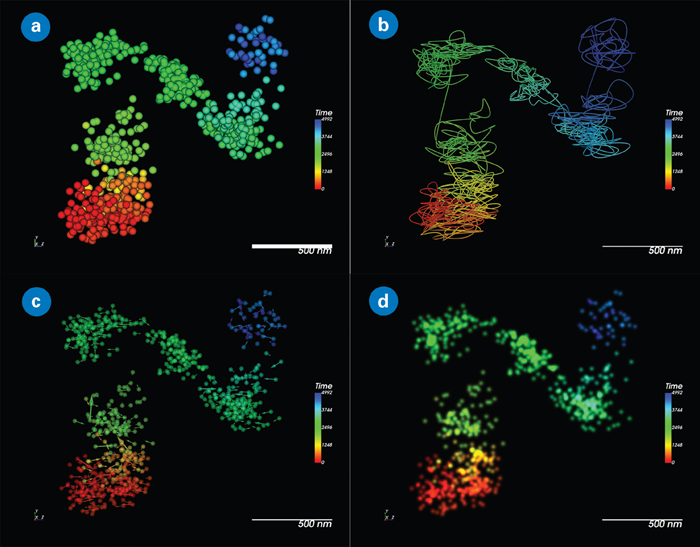
Figure 2. Static track visualization options in SRX. QD655 colored by time: (a) Point cloud of tracked localization; (b) Streamline representation; (c) Displacement vectors; and (d) Natural-image-like representation. Image Credit: Nano Surfaces and Metrology
Genetically encoded labels
Fluorescent Protein (FP) selection is necessary for single particle tracking. Brightness and photostability are the key considerations. With fluorescent proteins continually being developed, this list is bound to change. For now, the applications team suggests the options below.
When selecting an FP for a new line, strain, or plasmid, FPbase is a valuable resource, and the Bruker applications team can provide further assistance. It is also important to note that the labeled structure must be naturally sparse, with just a few spots present per cell at a given time, for fluorescent proteins to be tracked reliably.
Stably Fluorescent Protein Labels. Source: Bruker Nano Surfaces and Metrology
| Yello-Green (488) |
Orange (555) |
Red (647) |
| StayGold |
tdTomato |
tdSMURFP |
| mStayGold* |
mKate2 |
|
| mNeonGreen |
mScarlet* |
|
| meGFP* |
|
|
*Indicates monomeric
Halo and SNAP-tags
Halo or SNAP -tagging proteins provide versatility by allowing labelling first, with wavelength and technique selection determined later. There is also a broad range of specialized chemical dyes, many of which provide further advantages.
Options include Halo-functionalized self-blinking dyes and photoactivatable dyes for PALM and SPT-PALM. Halo and SNAP-tagged cell lines are commercially available, and strains/lines of model organisms may already be available for certain proteins of interest.
It is also possible to use photoconvertible fluorescent proteins for SPT-PALM, though they are generally less bright and exhibit less efficient photoconversion kinetics compared to chemical dyes.
Nevertheless, cell, tissue, and animal lines/strains may exist for different applications, as photoconvertible fluorescent proteins have been used extensively for time marking and versatility in developmental and cell biology. For SPT-PALM, HALO- or SNAP-tagged proteins and PA-JF dyes are generally recommended for visualization.
JF Dyes for PALM and SPT-PALM. Source: Bruker Nano Surfaces and Metrology
| Palm |
STP-PALM FPs |
| JF549 |
MEOS3.2 |
| JF646 |
Dendria |
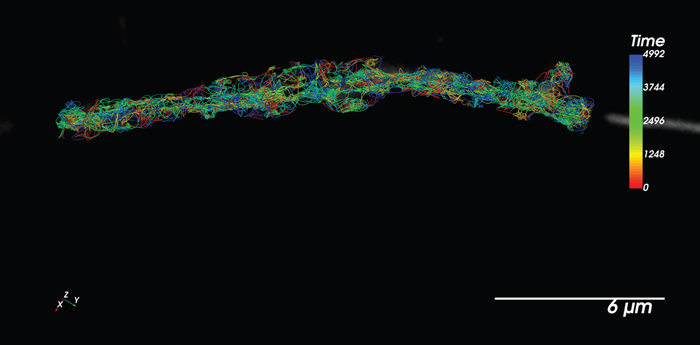
Figure 3. SPT-PALM using Halo-JF646. Particles tracked in concert with diffraction-limited imaging of organelles. Image Credit: Nano Surfaces and Metrology
Non-genetically encoded labels
In some cases, proteins, RNAs, biomolecules, or structures of interest can be directly or indirectly labeled with brightly fluorescent objects or dyes, then injected, transfected, or electroporated into the sample for subsequent study. Though low-throughput and potentially technically complex, this approach is excellent for single-particle dynamics studies.
Options include:
Quantum dots
- Inorganic, highly fluorescent structures
- Available functionalized
Gold nanourchins
- Inert spiky gold particles that emit light via surface plasmon resonance instead of fluorescence, and do not bleach
- Available functionalized
Primary antibodies or nanobodies
- Direct labeling of primary or secondary antibodies against the target biomolecule - suitable if the structure is externally accessible at the start of the experiment (e.g., receptor extracellular domains). Labeled primary antibodies and labeling kits are commercially available
Live RNA tracking
Several methods can be used to achieve live RNA tracking, all of which are compatible with the Vutara VXL.
- MS2/MCP (and related methods)
- Peppers (and related methods)
- Transfection of pre-labeled RNAs
Researchers can consult the relevant article or contact the applications team for comprehensive guidance on choosing techniques and labels for RNA imaging.
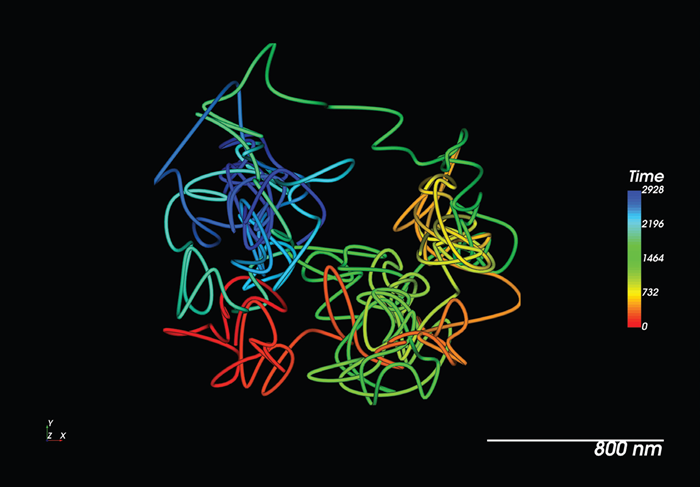
Figure 4. Live tracking of a single RNA molecule with MS2/ MCP in a living model organism. Image Credit: Nano Surfaces and Metrology
Single particle tracking with Vutara VXL
Effective single-particle tracking depends on careful label selection tailored to certain biological questions. Each technique carries distinct benefits and trade-offs.
Whether using the brightness and stability of quantum dots, the genetic precision of fluorescent proteins, or the versatility of Halo and SNAP tags, numerous factors must be considered and weighed against experimental goals. These factors include photostability, labeling density, and ease of implementation.
The distinct capabilities of the Vutara VXL super-resolution microscope and SRX software make this entire process both simpler and more precise, enabling advanced investigation of dynamic molecular processes.
About Bruker Nano Surfaces and Metrology

Bruker’s suite of fluorescence microscopy systems provides a full range of solutions for life science researchers. Their multiphoton imaging systems provide the imaging depth, speed and resolution required for intravital imaging applications, and their confocal systems enable cell biologists to study function and structure using live-cell imaging at speeds and durations previously not possible. Bruker’s super-resolution microscopes are setting new standards with quantitative single molecule localization that allows for the direct investigation of the molecular positions and distribution of proteins within the cellular environment. And their Luxendo light-sheet microscopes, are revolutionizing long-term studies in developmental biology and investigation of dynamic processes in cell culture and small animal models.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.