Bruker's Acquifer Imaging Machine (IM) is a fully automated widefield microscope with both brightfield and fluorescence imaging capabilities. It is best suited for high-content screening (HCS) and drug discovery experiments.
The Acquifer IM's static sample holding and movable optical unit keep samples stable throughout imaging. This is especially important for motion-sensitive samples like non-adherent cell cultures and embryos.
The system also offers a number of features that increase its utility and adaptability in automated drug discovery workflows, such as built-in temperature regulation, a robotic lid, and an open software interface.
This article describes how these distinct capabilities, along with the smooth implementation of automated processes, enable improved time-lapse investigations, screening, and high-throughput imaging tests.
Drug screening experiment design
Drug screening is a critical stage in medication development. It aids in the identification of prospective medicinal compounds, the evaluation of their efficacy, and the assessment of any side effects prior to clinical trials. Effective drug screening can also save time and resources by ensuring that only the most promising candidates advance.
An ideal setup for drug screening allows for straightforward drug administration. Drugs can be mixed into water for zebrafish or screened using 96-well plates for high-throughput analysis.
The Acquifer IM supports both of these experimental settings as it uses moving optics and excellent environmental control to preserve the samples. The Acquifer IM was designed in collaboration with active researchers to provide a suitable configuration for HCS drug screens using popular models such as cell cultures, organoids, and zebrafish.
Application example: Zebrafish HCS
In zebrafish HCS systems, efficiently managing multiple samples is critical. Typical zebrafish screens consist of hundreds to thousands of samples, with larvae investigated in bulk in 96-well microtiter plates. To accurately examine the phenotypic effects of medications, samples must be consistently aligned.
Random embryo orientation would make consistent, high-resolution imaging and capturing the same region of interest (ROI) difficult, both of which are critical for correct assessment.
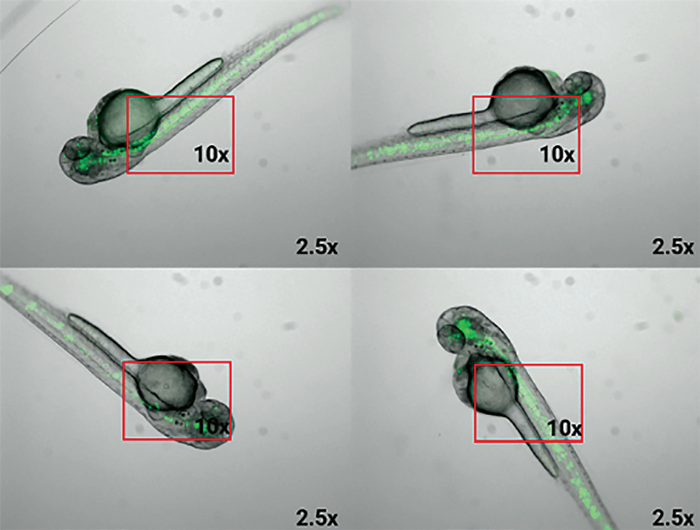
Figure 1. Random orientation in 2.5x magnification. Red rectangles show the tissue of interest’s field of view with a 10x magnification, and how random orientation would lead to data analysis challenges. Image Credit: Bruker Nano Surfaces and Metrology
To address this, scientists and Bruker's IM team collaborated on an experimental design that used adapters to make agarose gel indentations to consistently orient embryos in 96-well microtiter plates.
They used 3D-printed molds to form agarose gel in 96-well microtiter plates into a hollow, allowing the zebrafish larvae to align in the same direction. In addition, diverse geometries allowed the users to easily orient specimens dorsally or laterally.1
The Acquifer IM's sample-centered approach provides precision and dependability through friction-free linear motor technology and a motorized lid for robot-ready operation. Its moving optics keep the sample motionless, which is great for sensitive specimens, and built-in temperature adjustment ensures the best conditions.
Other capabilities include feedback microscopy, an open developer interface for scripting, and an optional photomanipulation module, making it an adaptable tool for sophisticated imaging requirements.
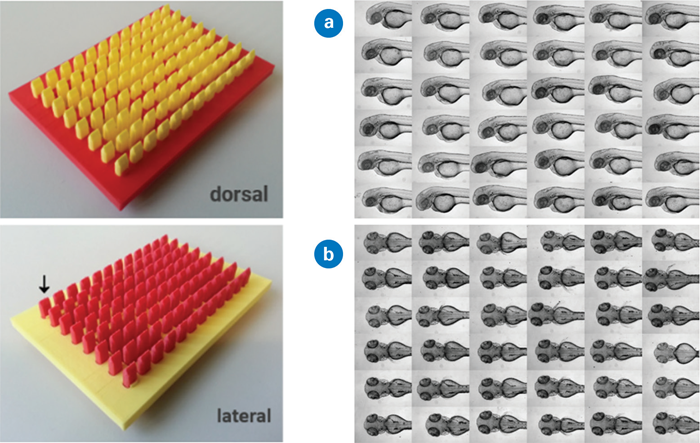
Figure 2. Left: 3D printed mold creates agarose gel indentation for standardized zebrafish embryo alignment. Right: Standardized embryo orientation with slightly different head positioning. Images used and adapted from [1] under a CC-BY 4.0 International License.
However, even with standardized body axis placement, embryo location may differ between samples, preventing standardized high-resolution imaging. To address this, the researchers developed Plate-Viewer visualization software, which allows for seamless labeling of regions of interest (e.g., tissues and organs) for future imaging.
This means that precise anatomical regions of interest can be obtained, and uniform data analysis is more simply accomplished. By standardizing the experimental setup with the Acquifer IM, 3D orientation tool, and Plate-Viewer software, data from complicated specimens may be gathered and analyzed in a high-throughput manner.
Application example: Automating workflows from sample preparation to results
To improve HCS experimental design, Westhoff et al. (2020) developed a methodology that included many semi-automated steps, such as targeted imaging of regions of interest, morphological characterization, and interactive data visualization.2
In this study, a compound library of authorized medications was used to screen for nephrotoxicity. The goal was to analyze 1280 chemicals in over 15,000 embryos and quantitatively evaluate 26 phenotypic changes. Such an initiative highlights the need for automated processes.
At 24 hours after fertilization (hpf), specimens were treated with chemicals from the Prestwick Chemical Library R© (Prestwick Chemical, D'Illkirch, France).
To standardize larval position, larvae were placed in an agarose gel fitted with 3D-printed molds at 48 hours post-fertilization. The Acquifer IM was then used to acquire a Z-stack of GFP and brightfield channels, with Z-stack centers determined using the GFP channel's built-in autofocus.
Brightfield images were used for broad phenotypic observations (malformation, oedema, etc.) caused by medication treatment, whilst GFP photos were manually annotated to precise kidney morphology levels. The study examined the effects of 1280 chemicals on over 15,000 embryos.
This resulted in 4.2 TB of data, which was stored and processed using Acquifer HIVE, Bruker's large data management system for centralized storage and computation.2
Automated imaging and processing
Semi-automatic ROI selection with the Acquifer Plate-Viewer
Semi-automated approaches to imaging and analysis are excellent for HCS since they involve minimal human interaction and no custom algorithm development. This strategy is useful when dealing with variable data that requires sophisticated image detection routines.
For supervised feedback microscopy, the Acquifer Plate-Viewer employs a semi-automated technique. It displays low-magnification pre-screen data from a whole microtiter plate in the plate layout, providing a quick and intuitive overview.
The integrated Click-Tool enables microscope users to select regions of interest for each well, while built-in Template Matching algorithms robustly pinpoint a wide range of target structures, including complex reporter expression patterns, morphological features, and unusual events.
Acquifer IM captures high-resolution data from specified locations depending on predefined settings, which are intuitively presented via the Acquifer Plate Viewer.
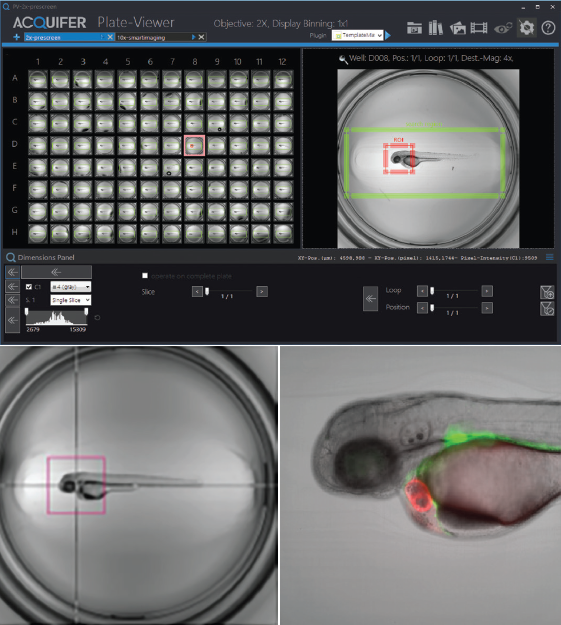
Figure 3. Illustration of Click-Tool functionality. Top: Three-day-old embryo of the epi:GFP;myl7mR transgenic line. Zebrafish embryos were visualized in Plate-Viewer software after the automated region of interest selection. Bottom left: The red bounding box indicates the field of view of a 10x objective used for subsequent high-resolution imaging. Bottom right: Single Z-plane of an automatically acquired high-resolution dataset. Image Credit: Nadia Mercader, Uni Bern.
Template matching
In HCS, automated object localization in biomedical imaging frequently depends on traditional image processing methods such as thresholding or edge detection. However, these methods frequently fail when images have low contrast or incomplete architecture.
As a result, more generic multi-step techniques can be effective for breaking down a large activity into manageable chunks.
Thomas and Gehrig previously presented a multi-template matching approach to boost detection capacity.4 The approach improved HCS and was implemented as a Fiji plugin, KNIME workflow, and Python package. It also improves object localization by leveraging numerous templates to facilitate automatic localization in microscope pictures.
This method's implementation, which does not require data pre-processing or annotation, is appropriate for a wide range of applications, including large-scale automated microscopy datasets and time-lapse analyses.
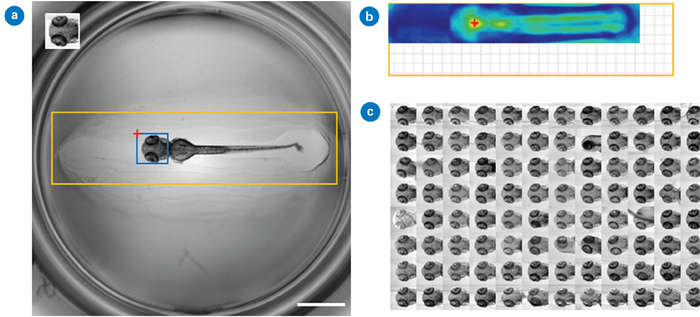
Figure 4. Detection of head regions in oriented zebrafish larvae using single template matching: (a) The searched image (2048 × 2048 pixels, scale bar: 1 mm) includes an inset template (188 × 194 pixels), a search region in orange (1820 × 452 pixels), and the predicted location in blue. The red cross marks the global maximum of the correlation map shown in (b); (b) Correlation map with the red cross indicating the bounding box position in (a). The grid area shows the correlation map's smaller size compared to the search image; and (c) Montage of detected head regions within a 96-well plate. Images used and adapted from [4] under a CC-BY 4.0 International License.
Batch processing with customized macro
In HCS, the ability to quantitatively analyze data is crucial, especially for tasks like morphological profiling after segmentation in phenotypic screening. Given the sheer volume of data involved, robust batch processing is not just helpful; it’s essential.
The Bruker ACQUIFER Fiji update site is a versatile toolbox for image processing, visualization, and batch analysis.5 It includes built-in tools like a plate montage viewer and supports image processing using Fiji Macros as well as other Fiji-compatible languages such as Java or Ruby.6
In addition, the updated site offers a wide range of examples and detailed documentation to support the development of analysis pipelines and smart microscopy workflows.
Application example: Understanding kidney development and disease from bench-to-bedside
Focal and segmental glomerulosclerosis (FSGS) is a set of diseases that scar the kidney's glomeruli, causing protein to seep into the urine. The absence of appropriate cell culture-based assays that reproduce physiological settings such as glomerular perfusion makes studying the causes and potential treatments of FSGS hard.
To circumvent this technical barrier, zebrafish can imitate kidney disorders using the nitroreductase/metronidazole (NTR/MTZ) system.7 The NTR/MTZ approach enables the induction of FSGS lesions by introducing medicines into the water, avoiding individual kidney injections.
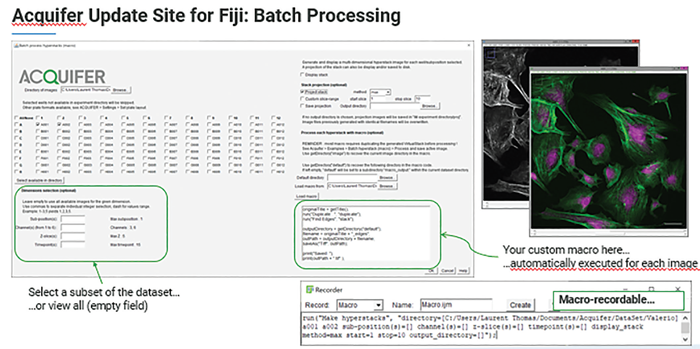
Figure 5. The Bruker Acquifer Fiji Update site is an ideal toolbox for image processing, visualization, and batch processing. Image Credit: Bruker Nano Surfaces and Metrology
Schindler et al. (2023) from the University of Medicine Greifswald used the NTR/MTZ system to produce FSGS lesions and screen a library of epigenetic modulators and FDA-approved medicines to see how they affected zebrafish kidneys.8,9
Acquifer HIVE was used to store and interpret thousands of zebrafish larval photos, allowing for automated analysis. Custom Fiji and image analysis macros enabled vascular segmentation and podocyte detection using mCherry fluorescence.
This high-content screening revealed that Belinostat is protective in an FSGS-like zebrafish model.
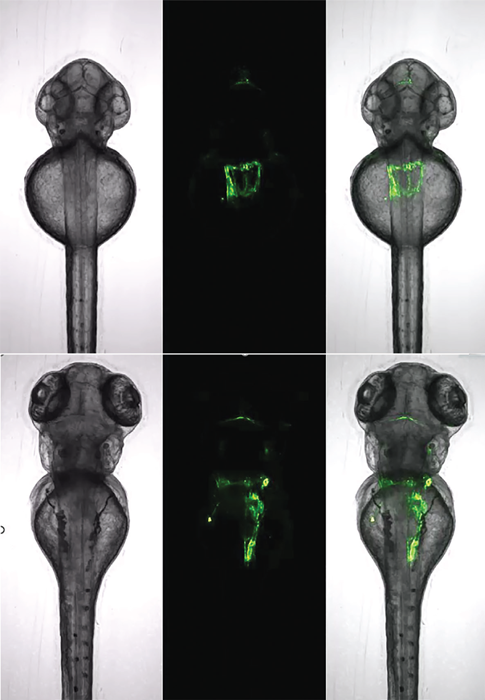
Figure 6. Zebrafish kidney analysis with a single dorsally oriented embryo from a 96-well plate acquired over a period of 29 hours (top: start, bottom: end). Tg(wt1b:EGFP) transgenic line. Images used from [1] under a CC-BY 4.0 International License.
Application example: Organoid development
In organoid research, drug screening has yielded promising results, for example, the use of a GSK3 inhibitor in Medaka-derived organoids. Inhibiting GSK3 activates the Wnt/β-catenin pathway, promoting the formation of retinal pigmented epithelium (RPE) marked by the specific expression of Otx2, an RPE marker.
This approach underscores the value of organoid models in drug discovery, offering insights into tissue-specific responses and the molecular mechanisms driving organogenesis.
Such advances highlight how organoid systems can be applied to high-throughput drug screening, accelerating the identification of therapeutic targets and deepening our understanding of developmental biology.
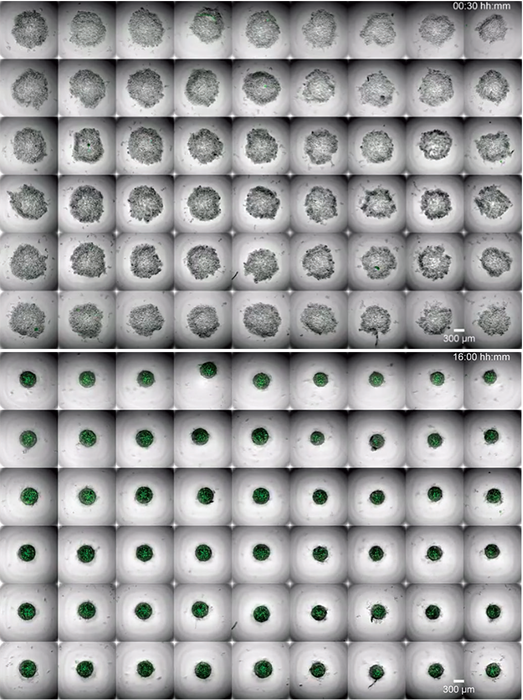
Figure 7. Still images from a video showing medaka blastula-derived aggregates undergoing morphological changes during the acquisition of retinal fate. Top: 00:30 hours; bottom: 16:00 hours. Images used from [10] under a CC-BY 4.0 International License.
Transforming drug discovery
Acquifer IM and Acquifer HIVE are well-suited for semi-automated HCS in drug discovery research. While the examples discussed here focus on zebrafish and organoids, the same workflows can be applied to other models, including cell cultures and patient-derived tissue biopsies.
By combining Acquifer IM’s automated widefield microscopy with Acquifer HIVE’s centralized data storage and processing, researchers can streamline their workflows, from sample preparation to final analysis, ensuring both efficiency and consistency.
Acknowledgments
This article is based on materials originally authored by Dr. Jochen Gehrig, Senior Product and Applications Specialist; Dr. Laurent Thomas, Software Engineer; and Dr. Elisabeth Kugler, External Marcom Specialist at Bruker.
References
- Wittbrodt, J.N., Liebel, U. and Gehrig, J. (2014). Generation of orientation tools for automated zebrafish screening assays using desktop 3D printing. BMC Biotechnology, 14(1). https://doi.org/10.1186/1472-6750-14-36.
- Westhoff, J.H., et al. (2020). In vivo High-Content Screening in Zebrafish for Developmental Nephrotoxicity of Approved Drugs. Frontiers in Cell and Developmental Biology, 8. https://doi.org/10.3389/fcell.2020.00583.
- Gehrig, J., Pandey, G. and Westhoff, J.H. (2018). Zebrafish as a Model for Drug Screening in Genetic Kidney Diseases. Frontiers in Pediatrics, 6. https://doi.org/10.3389/fped.2018.00183.
- Thomas, L.S.V. and Gehrig, J. (2020). Multi-template matching: a versatile tool for object-localization in microscopy images. BMC Bioinformatics, 21(1). https://doi.org/10.1186/s12859-020-3363-7.
- Schindelin, J., et al. (2012). Fiji: an open-source Platform for biological-image Analysis. Nature Methods, 9(7), pp.676–82. https://www.nature.com/articles/nmeth.2019.
- Acquifer (2022). ACQUIFER update site for Fiji. [online] YouTube. Available at: https://www.youtube.com/watch?v=eZ1cfOkkTEE [Accessed 24 Sep. 2025].
- Curado, S., Stainier, D.Y.R. and Anderson, R.M. (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nature Protocols, 3(6), pp.948–954. https://doi.org/10.1038/nprot.2008.58.
- Schindler, M., et al. (2023). A Novel High-Content Screening Assay Identified Belinostat as Protective in a FSGS - Like Zebrafish Model. Journal of The American Society of Nephrology, 34(12), pp.1977–1990. https://doi.org/10.1681/asn.0000000000000235.
- Schindler, M., et al. (2023). In vivo-Medikamenten-Screening zur Behandlung von Glomerulopathien. BIOspektrum, 29(5), pp.494–496. https://doi.org/10.1007/s12268-023-1997-5.
- Zilova, L., et al. (2021). Fish primary embryonic pluripotent cells assemble into retinal tissue mirroring in vivo early eye development. eLife, [online] 10, p.e66998. https://doi.org/10.7554/eLife.66998.
About Bruker Nano Surfaces and Metrology

Bruker’s suite of fluorescence microscopy systems provides a full range of solutions for life science researchers. Their multiphoton imaging systems provide the imaging depth, speed and resolution required for intravital imaging applications, and their confocal systems enable cell biologists to study function and structure using live-cell imaging at speeds and durations previously not possible. Bruker’s super-resolution microscopes are setting new standards with quantitative single molecule localization that allows for the direct investigation of the molecular positions and distribution of proteins within the cellular environment. And their Luxendo light-sheet microscopes, are revolutionizing long-term studies in developmental biology and investigation of dynamic processes in cell culture and small animal models.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.