Digestion is the process by which food breaks down into smaller compounds to allow the release and absorption of nutrients along the digestive tract. Studies have proven the significant impact of digestion on health, leading to the development of various types of in vitro digestion protocols that often yield non-comparable and inconsistent results.
In 2011, in response to these findings, the international INFOGEST network was established under a European Cooperation in Science and Technology (COST) Action [FA1005] to harmonize these protocols, enhancing the reproducibility and comparability of in vitro digestion studies.
This article presents a standardized approach for the application of the static INFOGEST protocol using the capabilities of BioXplorer 100.
The team’s findings revealed no significant differences in terms of lipid and protein digestion from an Ensure® Plus Vanilla when using the INFOGEST protocol in test tubes and the BioXplorer 100. This result demonstrates how automated systems can replicate the results of digestion simulations conducted in test tubes, showcasing the BioXplorer 100’s ability to lessen human error through automated design.
The continuous monitoring and correction of crucial parameters, such as pH and temperature, ensure that data is accurate and reproducible, supporting INFOGEST’s goal of standardized digestion simulations.
Overall, the BioXplorer 100 offers a reliable and efficient alternative to manual digestion methods, advancing the field’s ability to produce accurate and consistent results across different studies. This advancement promotes broader applications in in vitro digestion research, while enhancing understanding of digestive processes.
Introduction
Digestion is a complicated process involving the breakdown of food from large polymers, such as polysaccharides, lipids, and proteins, into smaller compounds.1 The aim of digestion is the release of nutrients from the food matrix, which in turn can be absorbed along the digestive tract.
Several phases are followed to achieve this: firstly, mastication and mixing with saliva in the mouth; after this, the movement to extremely acidic conditions in the stomach; and then the absorption of water, nutrients, and salt in the small and large intestines.2
In recent years, there has been an increased interest in comprehending the digestive process. Studies have shown that health and disease can be affected by the way food is digested.3 However, results regarding digestion simulation can be affected by the lack of homogeneity and standardization in protocols.4
To respond to these issues, INFOGEST was founded in 2011 as a European Cooperation in Science and Technology (COST) Action aiming to harmonize the in vitro protocols used by those in the scientific community who work on digestion simulation, improving the reproducibility and comparability of results across studies.
This collaborative effort has catalyzed the development of widely accepted in vitro protocols for both static and semi-dynamic approaches. In static models, physiological parameters, such as enzyme concentration and pH, are defined and maintained during the experiment: this approach is inexpensive and simple.9
This article demonstrates the use of H.E.L’s BioXplorer 100 to apply the static INFOGEST protocol.
Material and methods

Image Credit: H.E.L Group
The BioXplorer 100 is a multi-reactor system with eight zones that can be individually controlled, including agitation and temperature. In addition, pumps are connected to each reactor and can supply fluids. Each individual reactor was equipped with a pH and temperature process, sampling port, and four feeding lines. These lines provided simulated gastric fluids (SGF), an enzymatic mix, NaOH, or simulated intestinal fluids (SIF). The reactors were agitated with marine propellers. The system is controlled by H.E.L’s WinISO.
Solutions and enzymes
The digestion of lipids and proteins was tested using the convenient, liquid food Ensure® Plus Vanilla, which was acquired from Sorgente in the Netherlands and stored at room temperature until used, following its shelf-life recommendations. The composition of this drink was 20.20 % carbohydrates, 6.25 % protein, and 4.92 % lipids as per its label.9
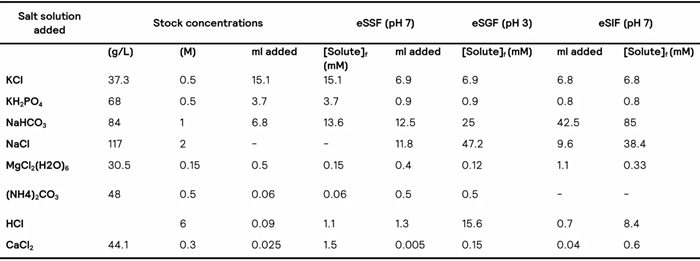
Table 1 shows the concentration of the different (stock) solutions to be used during in vitro simulations according to the Brodkorb et al. protocol.2
Table 1. Electrolyte solutions used in the digestion simulation. eSSF – Simulated Saliva Fluid, eSGF – Simulated Gastric Fluid, eSIF – Simulated Intestinal Fluid. Source: Taken from Brodkorb et al2
Porcine pepsin (3344 U mg-1), pancreas pancreatin (4xUSP) (amylase 41 U mg-1, lipase 36.3 U mg-1), trypsin (3.2 U mg-1), chymotrypsin (50.6 U mg-1) were acquired from Sigma Aldrich (Belgium).
Static conditions simulated
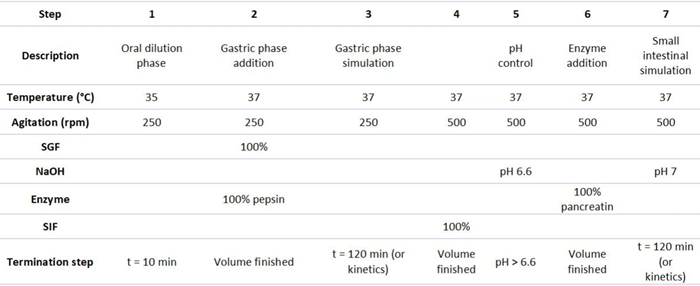
Table 2 demonstrates the physical parameters and the different solutions that were added during the simulation process. Step one covered the oral dilution and equilibration phase, performed for 10 minutes at 35 °C and 250 rpm. No digestive enzymes were added during this step, as mostly salivary amylase is of importance during oral simulation, but limited starch was present in the Ensure drink.
The rest of the steps were performed at 37 °C and 250 rpm. In step two (gastric phase fluid addition), the total SGF and pepsin volume were pumped into the vessel at the highest possible speed before proceeding to step three. SGF master mix was prepared according to the suggestions of Brodkorb et al. (2019) and included eSGF (pH 3), 0.3 M of CaCl2, 2 M of HCl, and milliQ water.
To determine the ratio of HCl to milliQ water added to the SGF mix, a preliminary experiment was conducted to reach a pH of three in the digest during the simulation of the gastric phase. Gastric lipase was not added at this stage since most lipids are digested in the intestinal phase.
In step three (gastric phase simulation), no solutions were added, but the temperature and agitation were maintained to allow substrate-enzyme interaction for maximally two hours. In step four, the SIF solution was added. SIF contained eSIF (pH 7), 0.3 M of CaCl2, and bile salts prepared in milliQ water (with a final concentration of 10 mM).
In step five, NaOH (2 M) was pumped to increase the pH to 6.6, which gradually increased during the following steps until a pH value of seven was attained during the simulation of the small intestine. In step six, the pancreatic enzyme solution was added using a cooled syringe pump, containing 100 U ml-1 trypsin, 25 U ml-1 chymotrypsin, 200 U ml-1 amylase, and 177 U ml-1 lipase. One of the pumps was equipped with a cooling mantle to maintain the enzymatic solution at four °C to preserve the enzyme activity throughout the simulation of digestion.
Finally, in step seven, the simulation of the small intestinal phase was performed. No solutions were added, but the temperature and agitation were kept consistent to allow substrate-enzyme interactions for a maximum of two hours.
Digestion was simulated in the available eight individual reactors. This allowed researchers to perform a kinetic digestion approach, meaning that the time-dependent digestion behavior could be analyzed from independent recipients. Based on insights gained from previous works, the following time points were chosen for evaluation in terms of digestion product formation: five, 10, 20, 30, 45, 60, 90, and 120 minutes after enzyme addition in the gastric or small intestinal phase.
When the gastric phase kinetics were evaluated, the pepsin activity was stopped by increasing the pH to eight at pre-determined moments. In the small intestinal phase, an aliquot of the digest was inhibited by adding 4-bromophenylboronic (one M in methanol) to inhibit pancreatic lipase, while another aliquot was placed in a water bath at 98 °C for five minutes to inhibit proteases and amylase. After inhibition, the digest (aliquots) were placed in an ice bath.
The lipid and protein content were quantified following the methodology throughout the simulation.
Table 2. Physicochemical parameters and solutions were added in the different steps of the digestion simulation. Source: H.E.L Group
Results and discussion
The accessibility of protein in Ensure® Plus Vanilla was evaluated using two approaches: a manual method with 10 ml tubes as described in the INFOGEST protocol, and an automated method using the BioXplorer 100.9 The manual tube method allowed a total volume of only 10 ml to be tested, whereas the BioXplorer 100 handled 80 ml.2
Two key differences between these approaches are their agitation and temperature control capabilities. In the tube method, samples were agitated on rotating wheels in a 37 °C incubator. However, the BioXplorer maintained a consistent temperature of 35 °C during the equilibration step and 37 °C during effective digestion simulation (remaining steps), with constant agitation at 250 rpm during the gastric phase and 500 rpm during the small intestinal phase.
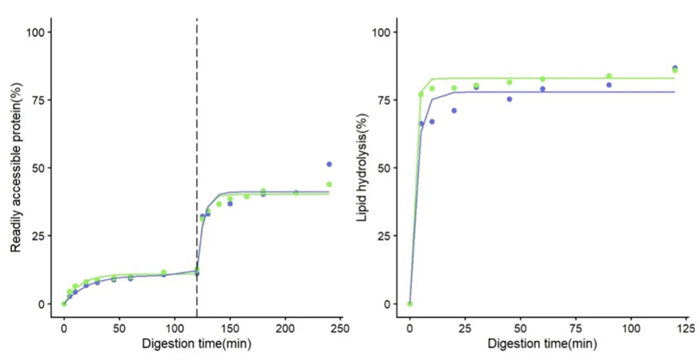
Figure 1. Evolution of the readily accessible protein and lipid in the simulation during the Gastric Phase (before 120 min) and Small Intestine phase (after 120 min). The dashed line represents the shift between phases. Colors represent the mean of testing (blue for tube, green for BioXplorer). Circles represent measured (circles) versus predicted values (solid line). Image Credit: H.E.L Group
Figure 1 illustrates the evolution of readily accessible protein and protein hydrolysis during the simulated process in tubes and the BioXplorer. Around 12 % of the protein was made available by the end of the gastric phase, reaching up to 44 to 51 % at the end of the small intestinal phase. Lipolysis was much faster, reaching values of over 60 %, only five minutes after the addition of pancreatic lipase.
No clear differences were seen between the results gained using the two methods. This suggests that the INFOGEST protocol can be implemented effectively using the BioXplorer 100. The automated processes in the BioXplorer minimize human handling, thus reducing potential errors. The ability to consistently monitor and adapt crucial parameters, including pH and temperature, ensures robust and reproducible data, aligning with INFOGEST’s goals to harmonize digestion simulations.
Conclusion
The results presented here underscore the efficacy of the BioXplorer 100 in implementing INFOGEST protocol. This system’s capabilities enable the implementation of automated control, offering a reliable and efficient alternative to manual methods.
Acknowledgments
Produced using materials originally authored by Sarah Verkempinck and Tara Grauwet from the Laboratory of Food Technology, and Dr. Mario Toubes-Rodrigo from H.E.L Group.
References
- Grundy, M.M.L., Moughan, P.J. and Wilde, P.J. (2024). Bioaccessibility and associated concepts: Need for a consensus. Trends in Food Science & Technology, 145, pp.104373–104373. DOI: 10.1016/j.tifs.2024.104373. https://www.sciencedirect.com/science/article/abs/pii/S0924224424000499.
- Brodkorb, A., et al. (2019). INFOGEST static in vitro simulation of gastrointestinal food digestion. Nature Protocols, 14(4), pp.991–1014. DOI: 10.1038/s41596-018-0119-1. https://www.nature.com/articles/s41596-018-0119-1.
- Ross, F.C., et al. (2024). The interplay between diet and the gut microbiome: implications for health and disease. Nature Reviews Microbiology, (online) 22, pp.671–686. DOI: 10.1038/s41579-024-01068-4. https://www.nature.com/articles/s41579-024-01068-4.
- Colombo, R., et al. (2021). Advances in static in vitro digestion models after the COST action Infogest consensus protocol. Food & Function. DOI: 10.1039/d1fo01089a. https://pubs.rsc.org/en/content/articlelanding/2021/fo/d1fo01089a.
- Zhou, H., Tan, Y. and David Julian McClements (2022). Applications of the INFOGEST In Vitro Digestion Model to Foods: A Review. Annual Review of Food Science and Technology, 14(1), pp.135–156. DOI: 10.1146/annurev-food-060721-012235. https://www.annualreviews.org/content/journals/10.1146/annurev-food-060721-012235.
- Menard, O., et al. (2023). Static in vitro digestion model adapted to the general older adult population: an INFOGEST international consensus. Food & function, 14(10), pp.4569–4582. DOI: 10.1039/d3fo00535f. https://pubs.rsc.org/en/content/articlelanding/2023/fo/d3fo00535f.
- Mulet-Cabero, A.-I., et al. (2020). A standardised semi-dynamic in vitro digestion method suitable for food – an international consensus. Food & Function, 11(2), pp.1702–1720. DOI: 10.1039/c9fo01293a. https://pubs.rsc.org/en/content/articlelanding/2020/fo/c9fo01293a.
- Minekus, M. et al. (2014) 'A standardised staticin vitrodigestion method suitable for food – an international consensus,' Food & Function, 5(6), pp. 1113–1124. DOI: 10.1039/c3fo60702j. https://pubs.rsc.org/en/content/articlelanding/2014/fo/c3fo60702j.
- S.H.E. Verkempinck, D. et al. (2022). Studying semi-dynamic digestion kinetics of food: Establishing a computer-controlled multireactor approach. Food Research International, 156, pp.111301–111301. DOI: 10.1016/j.foodres.2022.111301. https://www.sciencedirect.com/science/article/abs/pii/S0963996922003581.
About H.E.L Group
H.E.L develops and manufactures innovative scientific instruments and software designed to optimize the efficiency, safety, and productivity of key processes in chemistry and biology applications.
The H.E.L team of 70 includes highly skilled process and software engineers, based at their extensive research and manufacturing facilities in the UK, as well as sales and support offices around the world.
H.E.L has a long history of solving complex challenges for customers. Since 1987, the Company has worked with businesses and laboratories globally, providing proprietary automated solutions for the pharma, biotechnology, chemical, battery and petrochemical sectors.
We continue to extend the reach of our products and services to support and enable R&D and process optimization further across Europe, the US, China, and India.
H.E.L is accredited with ISO 9001 : 2015
Our mission
To help create a Healthier, Sustainable, Safer world for everyone.
Our vision
We equip scientists with the right tools and knowledge to develop safe, efficient new processes and molecules that benefit the world and its population.
Our values
- Insightful through experience. With over 30 years of in-house expertise and experience, we know how to overcome a challenge
- Collaborative by design. Dedicated to listening, learning, and working closely with industry experts, we empower others to fulfill potential
- Tenacious in spirit. Always looking for new and innovative solutions, we don’t stand still; instead, we are focused on reaching the next success
- Proud of progress. Fueled by our ability to make a real difference, while celebrating the achievements of others
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.