In this interview, Professor Matthias Mann discusses his work in the field of proteomics, and explains why applying proteomics-based technologies such as timsTOF to the clinic could accelerate the diagnosis and prediction of human disease.
What first attracted you to the field of proteomics?
I came from the mass spectrometry field before proteomics even existed. When proteomics came along, many people were focusing on the shape of 2D gels, but this didn’t interest me. Instead, the simultaneous analysis of many components in a cell and the systems biology aspects of shotgun proteomics appealed to me.
What sparked your interest in fundamental research while working in John Fenn’s lab?
I am educated as a physicist and a mathematician originally and started working with John Fenn on electrospray. I then followed the electrospray downstream, from characterizing single proteins to protein complexes, and then increasingly complex assemblies to whole cells, which I analyze today.
Following this, I began working on understanding how to create the ions, to how they can be measured, and then to increasing separation and speed to keep up with the complexity.
Rapid Proteome Analysis with timsTOF Pro - Professor Matthias Mann - Max Plank Institute
Why do you feel it's important to see more proteomics-based techniques in clinical environments?
Clinics rely heavily on laboratory tests, but it's usually one protein or one small molecule. The technical questions are about how accurate these are. For example, when faced with diabetics with high triglycerides, the laboratory scientists will know that the antibody will stop working, but the test doesn't say that. It could be made much more precise with mass spectroscopy-based techniques because they are a lot more digital.
The bigger reason to use mass spectrometry is that you can measure many things at once. I strongly believe that taking many measurements simultaneously, like a whole pathway or a whole assembly of proteins, would lead to much better diagnoses than what we have now.
Another aspect is that people are usually tested for one disease, but most people have co-morbidities which can be caught by using mass spectrometry in a systems biology-based approach.
Most often, the diseases will stem from the same basic cause. With diabetes, for example, the disease is caused by a metabolic syndrome which can manifest as diabetes, but also as liver disease and heart diseases. Taking a proteomics-based approach would force us to recognize that more than one thing is usually wrong.
 Yabusaka | Shutterstock
Yabusaka | Shutterstock
How useful might longitudinal health check-ups be for clinicians and patients?
This is a very attractive possibility and is not something that is done at present. Through our plasma proteomic profiling experiments, we discovered that people have stable levels of most of the proteins in blood plasma. However, we also found that although stable, these levels differ from person to person. It would, therefore, be useful to know what a person's baseline levels are before using a handful of these markers as a diagnostic indicator.
This would make the marker a lot more sensitive because we could see from the level we've established and see, for instance, that it has increased by 30%. In comparison, if we just had the population-based average, we would not know whether this 30% elevation is still in the normal range.
What is the timsTOF PRO and why is timsTOF, with specifically PASEF, an attractive solution for routine clinical applications?
The timsTOF Pro is attractive in many ways for us, particularly because of the extra dimension of the ion mobility. The ion mobility dimension is not new, but we've seen that we can use the tims dimension as an ion handling device. In effect, we use it to line up the ions and then do something with them one at a time as they elute from the tims device.
That's the basis for the PASEF; that we can sequence the ions one after another as they come out. In effect, we are sequencing them in parallel, thereby multiplying the sequencing speed. This is what makes the tims combination with time-of-flight (TOF) so attractive for data-independent acquisition.
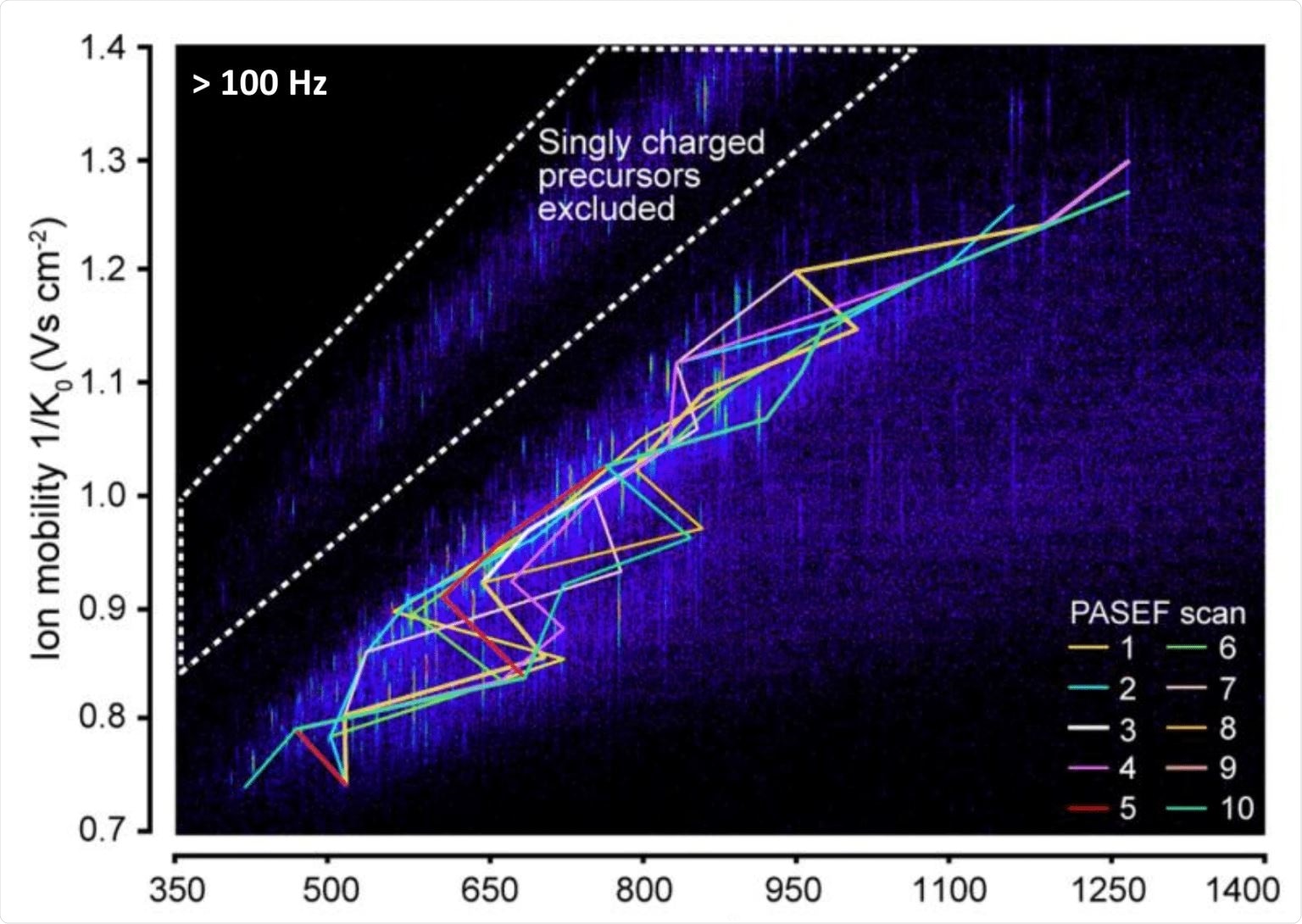 Credit: Meier, et al. Mol. Cell Proteomics. 2018.
Credit: Meier, et al. Mol. Cell Proteomics. 2018.
How do standard acquisition schemes compare to PASEF?
The advantage of PASEF is that you trap the ions once and then you can pick a lot of them for further manipulation, such as sequencing. This principle can be standard, for example, we know of twelve precursors per scan, but in the future, it might even be extendable to higher numbers. This would be possible by better engineering, pushing for more precursors, and driving things in that direction without affecting the sensitivity.
Standard acquisition schemes can be faster, but then inevitably, they become less sensitive because it’s done serially.
This means you trap or measure the precursor for a certain time and to increase the speed that time has to shrink together to get a higher frequency. That would always be at the expense of sensitivity, which is not the case for PASEF.
In which clinical fields would you like to see PASEF data being used and why?
Two big directions are measuring blood tests and tissues. We're very attracted to measuring the plasma proteome because that's minimally invasive and only needs a drop of blood that you can get from any person without issue. For tissues, it's more important for serious conditions like cancer, which is more tissue-dependent, as opposed to the plasma proteome, which can be done for a lot of people that aren’t necessarily sick.
I would like to PASEF being used for at-risk populations, for example, to diagnose whether they're at risk for liver disease. We could also look at those already diagnosed with liver disease and then test them again after an intervention. This requires a lot of frequent blood tests, which would therefore benefit from being fast and cheap which I think is within the remit of the technology.
To do this, we'd like to combine the Bruker timsTOF with the Evosep which has a very fast gradient so we can do 60 segments per day. That alone is good utilization of the mass spectrometer, which can help justify the costs. Even in the short run, you could do this analysis for a couple of hundred dollars or euros and in the long run it could be less than a hundred euros.
If you compare this to ELISA tests, which can cost forty dollars and are less sensitive, the answer is simple. We need to work on the dynamic range, so the antibodies can go deeper down in the dynamic range of the plasma proteome.
 angellodeco | Shutterstock
angellodeco | Shutterstock
Do you think depletion could be a solution?
Yes, definitely. With the way they're done now it wouldn’t be possible, because right now you need very expensive columns and they degrade over time which isn’t good for clinical applications. However, in some shape or form, depletion will be part of the solution.
As an example, what can PASEF data reveal about cancer cells?
We currently use PASEF for pathology samples, by taking FFPE slices and cutting out affected areas with laser microdissection. Cancers are highly heterogeneous, so we cut out the more advanced areas of the cancer from the less advanced and then we compare them.
By comparing the proteome, we can see what's different, which will give us extra targets. We use advanced software for this to look at what is upregulated in this cancer and which of these proteins correspond to drugs that are available. We've have done this already from a point of care or for patients that were chemorefractory, and we have found new targets that allowed them to receive further highly targeted medication.
This approach is not approved for widespread use, but we are still working on this project with small patient cohorts. The patient has to approve, the doctors have to approve, and currently, this is purely for cases where every other therapy has been exhausted.
How can PASEF data be used to inform and personalize treatment plans for patients with metabolic syndrome?
We can see proteins that inform on the inflammatory status of the patient and then take that panel, with other panels too, to inform about certain aspects of the health state of that individual. Then we would be able to say that this person has low-grade inflammation due to being obese, for instance, and the doctor could suggest a number of lifestyle interventions, such as exercising more or losing weight.
However, some people may benefit more from exercise and another person may benefit more from losing weight. This is well-known in practice, but there's no readout for that so far when the doctor has to make a recommendation. We can use our technology to get these inflammatory panels and say the exercise without losing weight is enough to decrease the low-grade inflammatory state.
How do you think machine learning will enhance the capabilities of timsTOF, and aid its use in a clinical setting?
We're always excited about advances that are being made in machine learning and deep learning and how to apply them in our analytical practice. These technologies come in at two levels. One level is that they can predict the extra measurements. For example, we've done work on measuring the law of collision on over 2 million cross-sections and then we can use a deep-learning framework to accurately predict what the cross-section will be for a given peptide sequence.
The second level is to predict the retention time. Other people have shown that this can be done MS-MS spectra, too. This allows us to predict the peptide sequence, what its behavior will be, or what its position in the multi-dimensional data cuboid will be. This would help us to identify the peptides.
Going further downstream we can use machine learning to classify the patients, based on their proteome, into risk states. This removes the need for a single biomarker and instead allows you to use the whole pattern of protein expression to see whether there is a risk of disease. More data may allow us to apply deep learning in this context within a year or two.
 Javier Regueir | Shutterstock
Javier Regueir | Shutterstock
What does the future hold for your research?
We've focused a lot on clinical proteomics, but our lab also studies many other things like signaling analysis or phosphorylation. That said, I think my group can make the biggest impact in clinical proteomics with the plasma proteome because this could be used so generically as a diagnostic for many diseases.
Our goal is to develop the research further, to a state where it can be used to diagnose patients with a number of human diseases. We want to use this as a stepping stone to start predicting the risk of other diseases such as metabolic syndromes, cancer (including late-presenting cancers), and completely different diseases such as psychiatric or neurodegenerative diseases.
Watch the presentation
Please note: Although there is a high potential for use in a clinical setting (patient screening and/or diagnosis), these research instruments and workflows have currently not been validated or approved for clinical diagnostic use.
ASBMB 2019 Presentation - Matthias Mann
Where can readers find more information?
About Professor Matthias Mann
 Professor Matthias Mann is the head of the Department of Proteomics and Signal Transduction at the Max Planck Institute of Biochemistry. He is one of the most cited researchers in the world, with over 200,000 citations. The Mann lab has been leading the advancement of mass spectrometry-based proteomics for the past two decades.
Professor Matthias Mann is the head of the Department of Proteomics and Signal Transduction at the Max Planck Institute of Biochemistry. He is one of the most cited researchers in the world, with over 200,000 citations. The Mann lab has been leading the advancement of mass spectrometry-based proteomics for the past two decades.
Professor Mann is also the Director of the Proteomics Program at the Novo Nordisk Foundation Center for Protein Research (NNF-CPR) in Copenhagen, where he leads the Clinical Proteomics Group.
The group has worked on a large number of international projects including the MSmed EU project, the ERC Synergy Grant ToPAG, Michael J. Fox Foundation for Parkinson’s Research projects, and Digimed Bayern.