This article and associated images are based on a poster originally authored by Alex Chenchik, Lester Kobzik, Tianbing Liu, Dongfang Hu, Kitt Paraiso, Khadija Ghias, and Paul Diehl and presented at ELRIG Drug Discovery 2025 in affiliation with Cellecta, Inc.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
Adaptive immune receptor (AIR) sequencing of T- and B-cell receptors (TCRs, BCRs) has broad applications in infectious disease, autoimmunity, and cancer immunotherapy, but connecting receptor discovery to antigen specificity remains a major bottleneck. The researchers present a comprehensive workflow that integrates bulk and single-cell repertoire sequencing, paired-chain reconstruction, and functional assays to bridge this gap. By sequencing DNA and RNA from the same sample, the study distinguishes clonal expansion from transcriptional activation.
In the work, paired TCR sequences are resolved using a plate-based, single-cell assay, cloned, and expressed in GFP-reporter Jurkat cells. These engineered cells enable sensitive antigen-screening assays, including dextramer binding and co-culture with K562 antigen-presenting cells, yielding fluorescent readouts of antigen recognition.
This platform unifies discovery and validation, providing a direct route from repertoire profiling to antigen identification. It can be applied to map disease-relevant clonotypes, interrogate peptide and small-molecule libraries, validate immunotherapies, and dissect receptor signaling pathways that underlie adaptive immune responses.
Introduction
- The adaptive immune system is responsible for the antigen-specific immune response driven by the lymphocytes (B-cells and T-cells) through cell surface receptors, resulting in immunological memory.
- Genetic recombination of V, D, J, and C regions in T- and B-cells generates millions of different T-cell receptors (TCR) and B-cell receptors (BCR), creating a highly complex “universal response” to almost any pathogen.
- Understanding the complex interactions between disease and the immune system enables the discovery of diagnostic, predictive, and prognostic biomarkers and therapeutic targets.
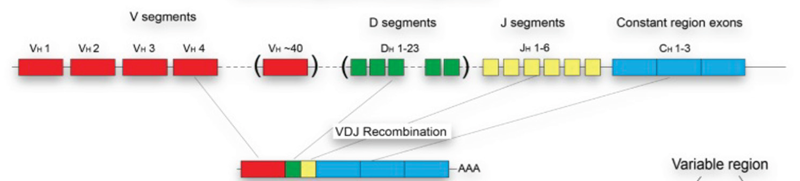
Figure 1. Schematic representation of TCR repertoire diversity and VDJ recombination. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Method
We demonstrate an integrated workflow that focuses on three analytical approaches, which together provide a comprehensive approach to identify and characterize therapeutically important TCR/BCR-antigen interactions.
Objectives of immune repertoire studies
- Repertoire diversity analysis to assess diversity and identify activated clonotypes, typically from bulk cell samples.
- Matching chain-pair sequences (TCR-α/β, BCR-heavy/light) to characterize whole active receptors, typically by analysis of selected pools or single cells.
- Identifying antigen-receptor interactions to find effective receptors and predict antigen targets based on receptor sequences.
Repertoire diversity analysis
Repertoire diversity was analyzed using the DriverMap™ Targeted Multiplex RT-PCR technology, which enables simultaneous amplification and sequencing of 100,000 TCR and BCR receptors in a single assay. Empirically tested optimized primers enable the following:
- Profiling of either the CDR3 or full-length CDR1-CDR2-CDR3 regions from DNA (CDR3-only) or RNA samples
- Unbiased amplification without primer-dimers and off-target products
- Quantitative clonotype analysis
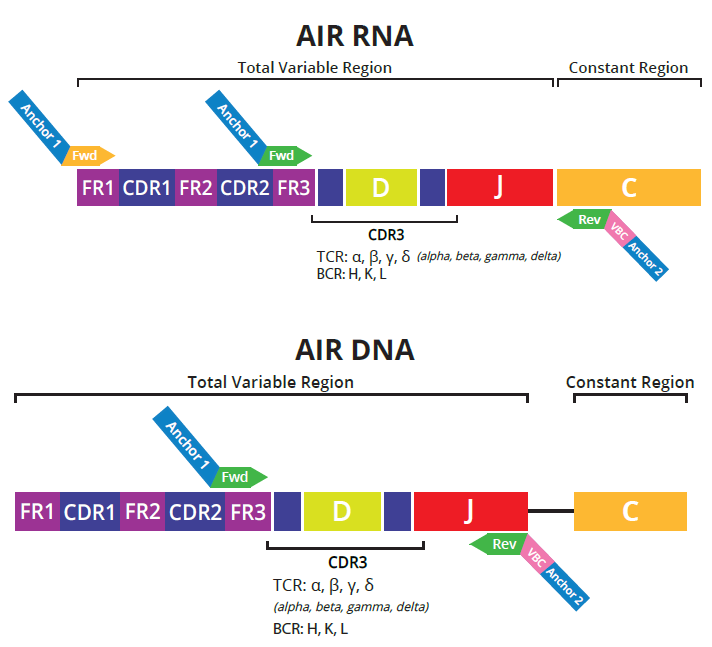
Figure 2. Schematic for multiplex RT-PCR primers for RNA and DNA assays. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Key benefits
- Flexible input: Accurate, reproducible profiling from DNA or RNA.
- Optimized for challenging samples, including FFPE, tumor biopsies, and microsamples (e.g., as little as 30 μL of blood).
- CellDirect Protocol enables direct amplification from cell lysates for a small number of cells without requiring RNA purification or enrichment.
Combined RNA and DNA profiling
Tumor-activated clones
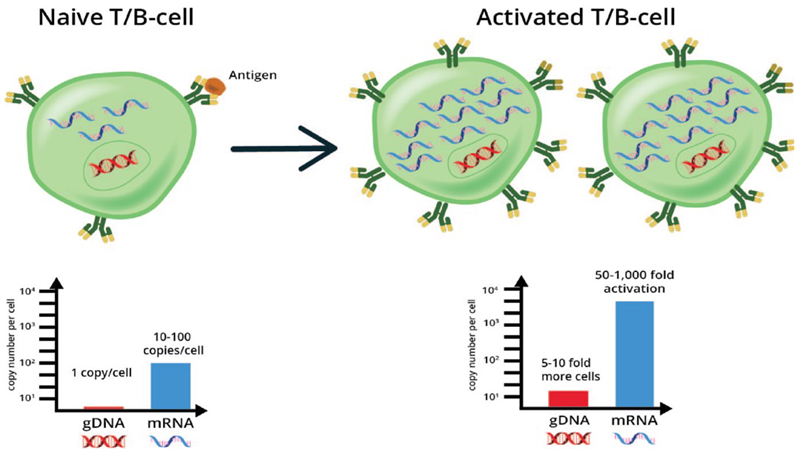
Figure 3. Schematic shows how the level of receptor RNA and DNA before and after antigen activation can be used to differentiate clone proliferation from increased receptor expression. Parallel profiling and normalization of RNA and DNA can elucidate differences between induced expression (i.e., gene activation by RNA assay) from proliferating cells clones (by DNA assay). Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Vaccine-induced immune receptors
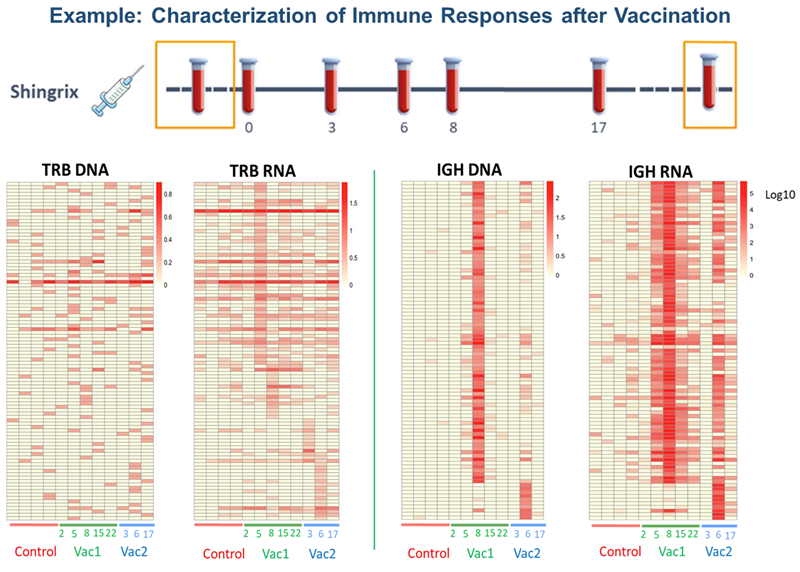
Figure 4. Peripheral blood was collected for controls (pre-vaccination and multiple weeks after vaccination) and treatment (few days/weeks post-vaccination). Longitudinal analysis of the adaptive immune repertoire by DriverMap™ AIR RNA and DNA shows differences in sensitivity in identifying clonotype activation pre- and post-vaccination. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Combined RNA and DNA profiling
- Samples from patients with T-cell large granular lymphocytic leukemias (T-LGL) at three different timepoints (3 plates) were analyzed using DriverMap™ Single-Cell AIR Immunophenotyping Kit.
- Results revealed a highly abundant TCR with specific chain-pairs in a T-LGL patient with severe neutropenia.
- Database (VDJdb server) analysis identified putative antigens recognized by the identified TCR.
- The most abundant receptor was cloned and expressed in Jurkat cells and then tested against predicted candidate antigens to confirm the target.
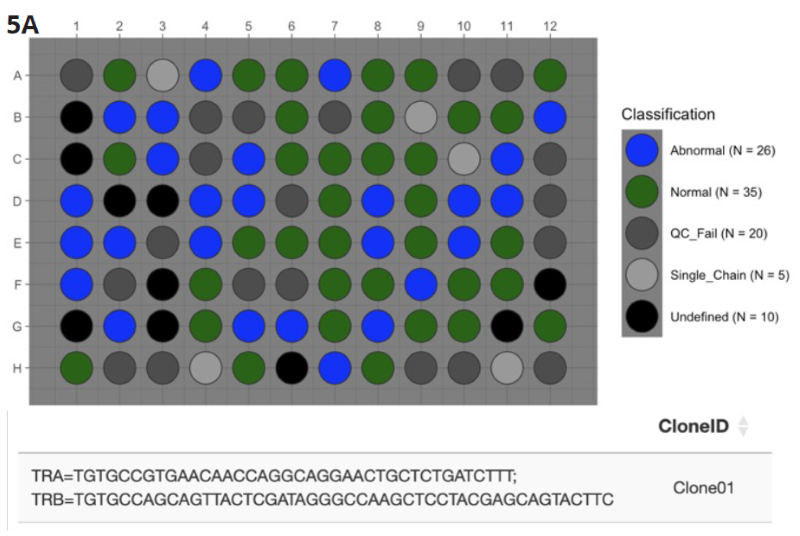
Figure 5A. DriverMap™ Single-Cell AIR Immunophenotyping Kit which, in addition to profiling TCRs, includes multiplex primers in the same reaction to profile expression of 36 T-cell marker genes, to identify the most abundant single TCR α/β clonotype pair in leukemic T-cells (35 “Normal” blue wells). Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
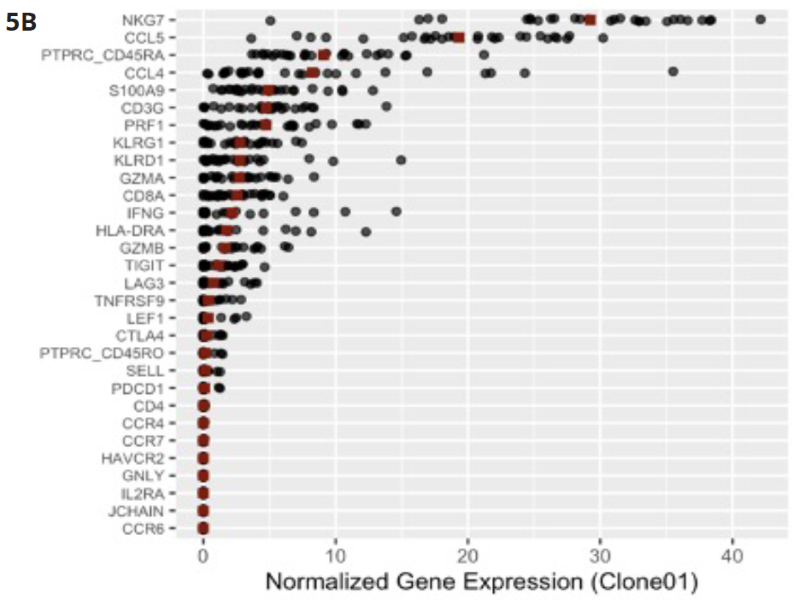
Figure 5B. Gene expression analysis of the primers for the 36 T-cell marker genes included in the assay revealed that the NKG7 and CCL5 are highly expressed in the most abundant clone. Co-expression of these genes has been associated with neutropenia. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Decode TCR-antigen interaction
By expressing a TCR-αβ receptor in Cellecta’s Jurkat TCR KO GFP Reporter cells, it is possible to test and screen TCRs with multiple antigens to identify which ones they react with.
Jurkat TCR-antigen reporter cells & matched peptide-dextramer
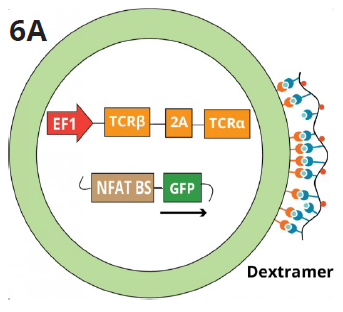
Figure 6A. The schematic shows the Jurkat TCR K/O NFAT-GFP reporter line transduced with a control TCR construct targeting EBV, FLU, or CMV peptides. Fluorescently labeled dextramers specific for each peptide bind to the corresponding TCRs expressed on the surface of the Jurkat reporter cells and detected via FACS analysis. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
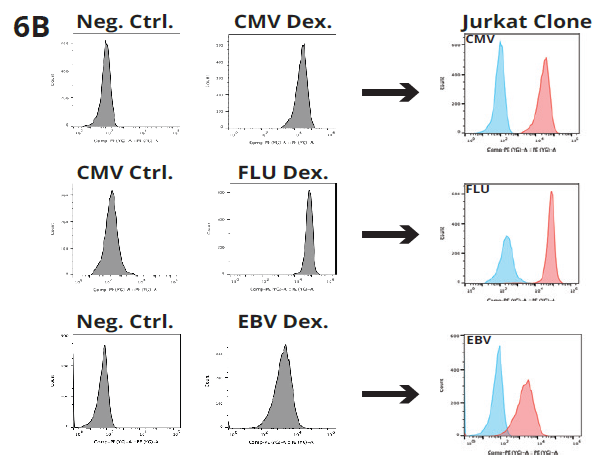
Figure 6B. Jurkat Reporter Clone staining with peptide-specific dextramer (CMV, FLU and EBV). FACS results show positive staining of Jurkat clone expressing peptide-specific TCR. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Jurkat TCR-antigen reporter cells and K562 APC co-culture
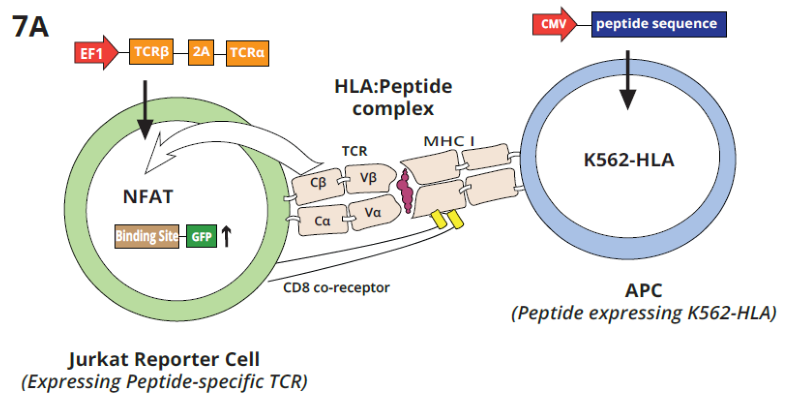
Figure 7A. Schematic shows the interaction of a Jurkat GFP reporter cell line expressing a specific TCR with K562 APC cells expressing the complementary antigen. TCR-Epitope-MHC interactions activate expression of GFP, which labels APC-interacting peptides. Amplification and NGS analysis from APCs allow the identification of peptide epitope candidates. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
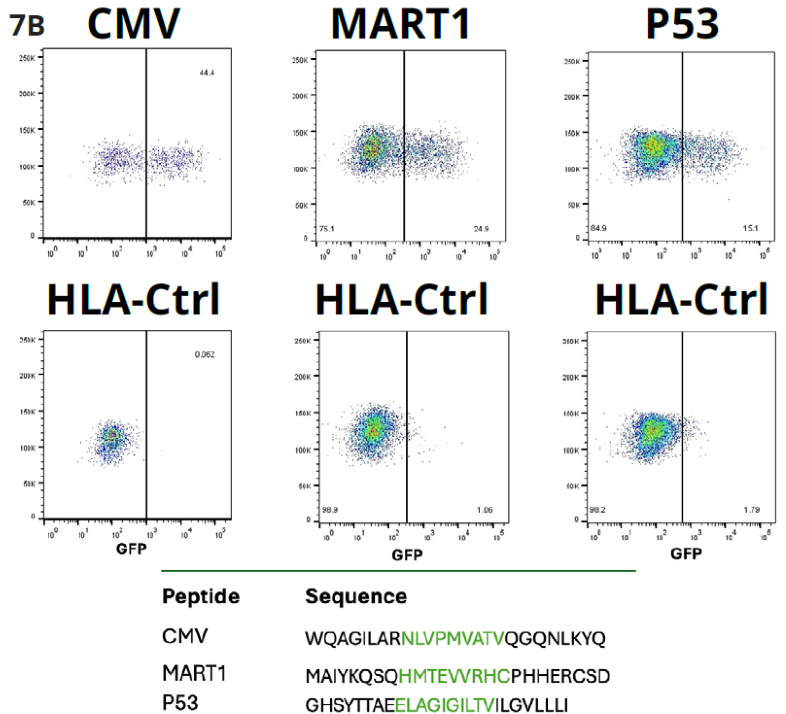
Figure 7B. Shows that the Jurkat NFAT GFP reporter cells expressing CMV, MART1, or P53 peptide-specific TCR and K562_HLA cells expressing CMV, MART1, or P53 short peptide sequence were co-cultured with each peptide-specific TCR reporter cell (Jc/Jm/Jp). FACS results show activation of the Jurkat reporter cells after incubation as compared to the negative control. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Discussion
Cellecta provides a comprehensive suite of products and services for bulk and single-cell immune profiling, cloning, and packaging, as well as Jurkat and APC reporter cell lines, enabling the efficient characterization of T-cell or B-cell receptors for biomarker and therapeutic discovery and validation.
- Step 1: Screening activated clonotypes - DNA and RNA are analyzed in parallel, enabling the identification of both activated receptors (RNA) and clonally expanding T- or B-cells (DNA), providing a comprehensive view of immune activation and clonal proliferation.
- Step 2: Identifying receptor chain pairs: essential to characterize the structure of the functional TCR or BCR molecule and enable testing or screening for antigen responses. We have demonstrated how these chain pairs can be readily identified starting from single-cells or focused cell populations.
- Step 3: Screening TCR-antigen interactions: Once the structure of the whole receptor is clear, it can be cloned into a Jurkat-TCR knockout line where it is expressed and can be used as a reporter line, in conjunction with modified antigen-presenting K562 cells or a dextramer-conjugated peptide.
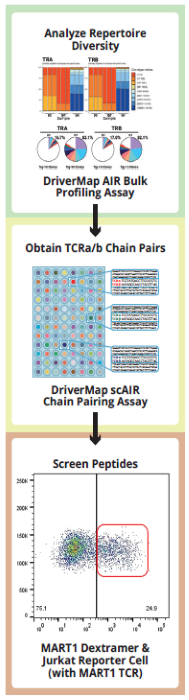
Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
About Cellecta
Cellecta, Inc. was founded in April 2006 by one of the chief scientists from Clontech Laboratories. The company was borne out of the need for higher quality, more advanced shRNA and other lentiviral libraries. Our goal was to develop advanced high-throughput (HT) genetic screen technologies and their applications for the discovery and functional characterization of novel therapeutic targets and drugs.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 26, 2025