This article and associated images are based on a poster originally authored by Alex Chenchik, Tianbing Liu, Dongfang Hu, Kitt Paraiso, Lester Kobzik, Khadija Ghias, and Paul Diehl and presented at ELRIG Drug Discovery 2025 in affiliation with Cellecta, Inc.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
This study presents a cost-effective method for single-cell immune receptor (TCR/BCR) profiling using a standard 96-well plate, requiring no specialized equipment like microfluidic platforms. T- or B-cells are flow-sorted into wells, then primers for immune receptor and key marker genes are added, followed by multiplex RT-PCR and next-generation sequencing. Validator barcodes (VBCs) ensure robust clonotype quantification with minimal cross-well contamination. Data analysis yields full-length receptor chain-pairing, clonotype abundance, and cell subtype information via gene expression profiles. This streamlined assay – ideal for rare or enriched cell populations of a few thousand cells – provides comprehensive single-cell adaptive immune receptor profiling, and enables accurate detection of antigen-activated lymphocytes without costly reagents or instrumentation. The approach will facilitate translational applications such as engineering T-cell-based cancer therapies or therapeutic antibodies for immunotherapy of cancer and autoimmune diseases.
Introduction
- Hundreds of millions of different T- and B-cells with unique TCRs and BCRs define differences in our immune responses.
- Sequencing of these diverse repertoires is called adaptive immune receptor (AIR) profiling.
- Single-cell AIR profiling allows one to obtain chain-pairing information and a gene expression profile of each cell type.
- However, single-cell assays are complicated to run, require expensive reagents and have limited sequencing throughput with the standard single-cell platforms.
- Cellecta offers a 96-well, plate-based scAIR sequencing assay, available as a kit or service. Obtain complete coding region sequences for human, mouse, and mouse/human hybrid T-cell (TRA-TRB) or B-cell (IGH-IGK-IGL) receptors.
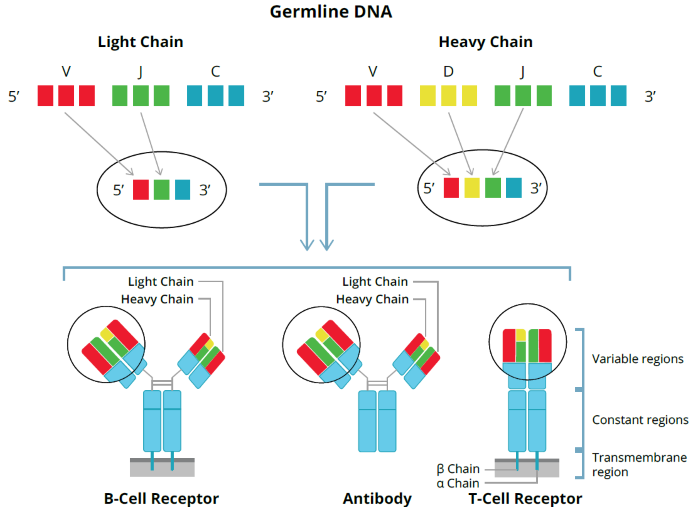
Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Method
- Sort cells into an indexed 96-well plate with RT primers, hybridize, then pool samples into a single reaction tube. Perform RT-PCR and next-generation sequencing (NGS), scTCR-Seq or scBCR-Seq. No RNA extraction, specialized equipment, or complex barcoding required.
- Two workflow options--the first for 96 flow-sorted individual cells per plate; and the second for up to ~10,000 cells (~100 cells/well) to thoroughly characterize each receptor in small numbers of cells, or extract large numbers of abundant receptor chain pairs in larger cell populations.
- An integrated bioinformatics analysis pipeline with MiXCR software on Platforma (MiLaboratories, Inc.) is used to process FASTQ files, obtain clonotype alignment and chain-pair information, and, optionally, for human samples, analyze expression of key marker genes that show the immunophenotype of each cell. Other open-source tools may also be used for further data processing.
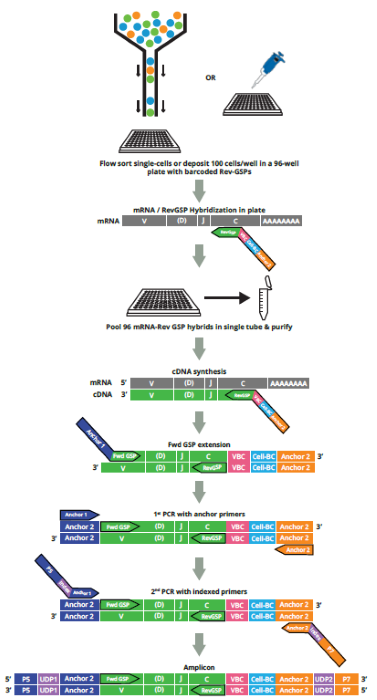
Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
- RT-PCR-based multiplex assay for TCR α/β or BCR (IGH, IGK, and IGL) primer pairs with crucial T/B-cell marker genes for parallel immunophenotyping.
- Improved coverage of CDR1-CDR2-CDR3 (full-length) regions with highly validated, redundant V primer sets.
- Unbiased amplification with universal anchor primers.
- No primer-dimers and off-target products.
- Quantitative clonotype analysis with VBCs and AIR RNA calibration standards.
Single-cell AIR validation
Plate uniformity test
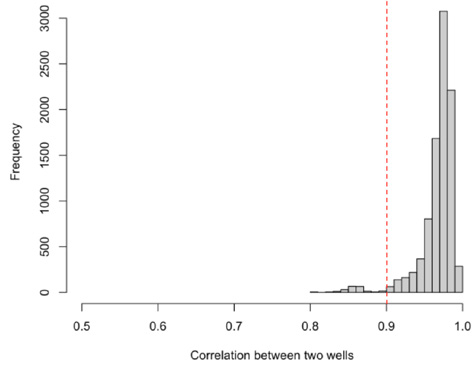
Fig 1. Conducting a plate uniformity test shows >90 % of correlation between ‘normal’ wells with a low SD. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Contamination test
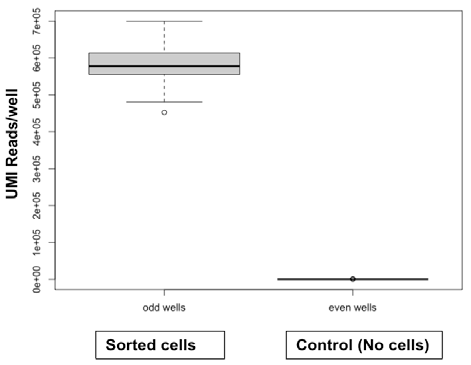
Fig 2. Cross-contamination test between odd wells with sorted cells and control even wells without sorted cells shows no cross-contamination. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Case study I: Detection of CMV-TCR clones
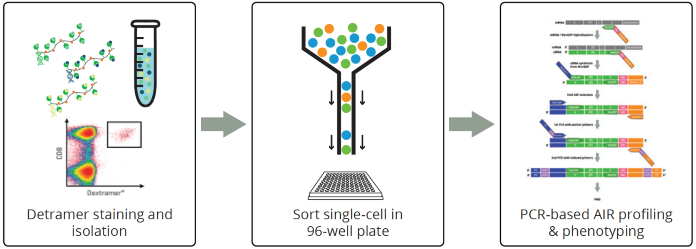
Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
- Proof-of-principle experiment was conducted in CMVpositive, PBMC stained single-cells with CMV-specific MHCpeptide Dextramer Reagent (Immudex) sorted in 96-well plate.
- As a control, we sorted non-stained single cells (negative control) and PHA-activated single cells (positive control) into a 96-well plate.
- Separate scAIR-TCR-Mark36 assay amplification and sequencing was performed and run through MiXCR pipeline.
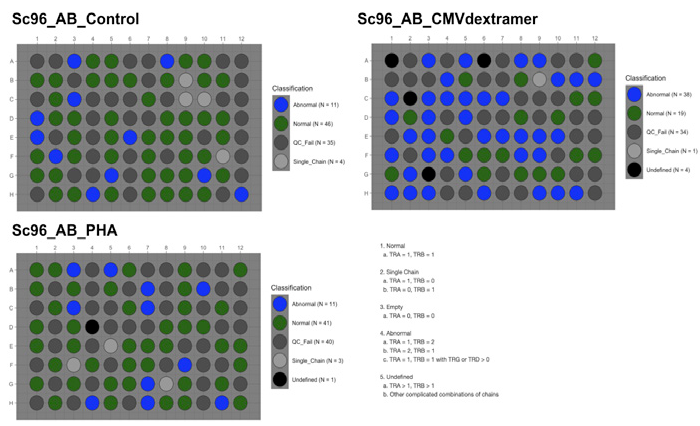
Fig 3. Classification of wells indicates the number of Normal (single-chain) wells obtained from the sample and control plates. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
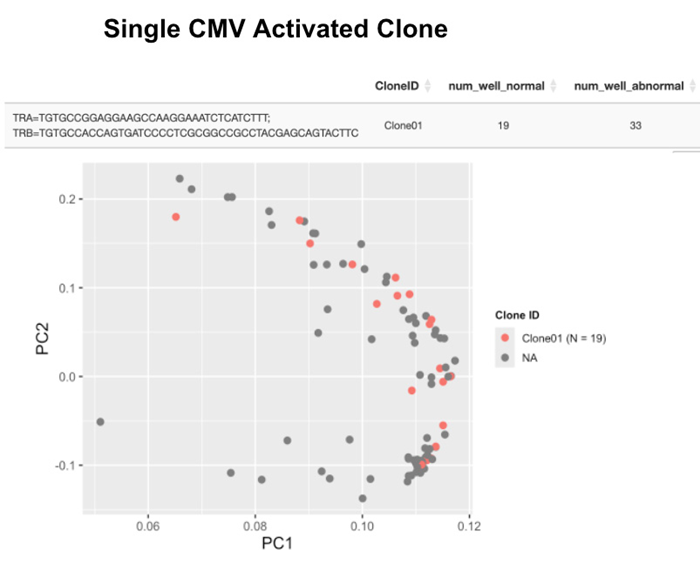
Fig 4. A single CMV-specific clone was isolated from a CMV Dextramer-stained sample, representing CD8+ cytotoxic cells and an activated memory cell phenotype characterized by high expression of effector genes, including NKG7, CCL5, GZMA, PRF1, and IFNG. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Case study II: Single-clone T-cell leukemia
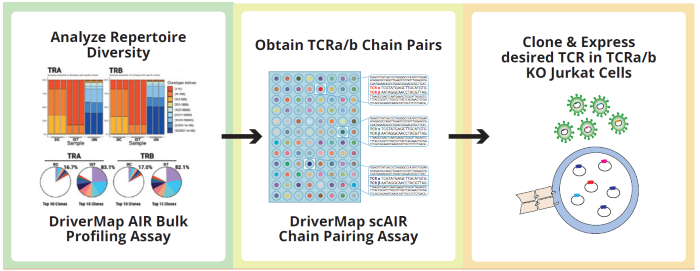
Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
- Patient samples with T-cell large granular lymphocytic leukemias (T-LGL) at three different timepoints (3 plates) were analyzed using Cellecta’s single-cell AIR-TCR-Mark36 profiling assay.
- Single-cell AIR profiling revealed the most abundant TCR clonotype chain-pairs in a T-LGL patient with severe neutropenia.
- Further analysis of the CDR3 region (VDJdb server) identified putative antigens recognized by the expanded clonotypes.
- The most abundant full-length receptor clonotypes were cloned and expressed in Jurkat cell lines and screened against candidate antigens to devise a model for the onset and development of T-LGL.
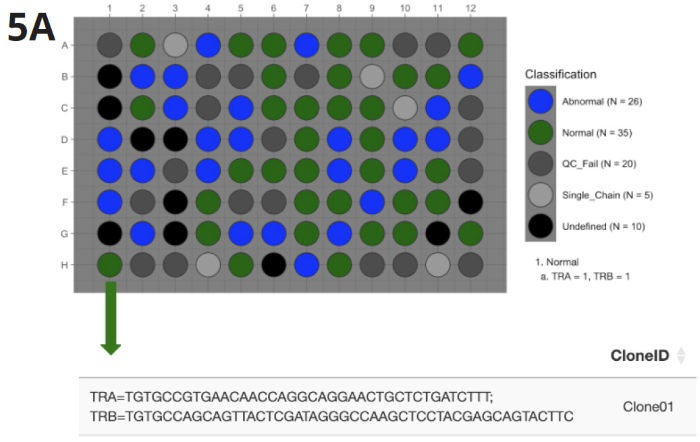
Fig 5A (top). Most abundant single TCR α/β clonotype pair was identified in 35 wells (Normal) across all three time points. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
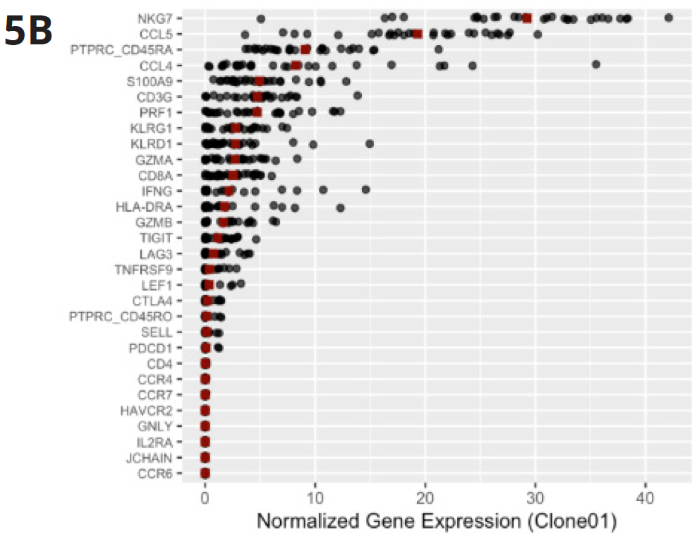
Fig 5B (bottom). Gene expression profiling analysis revealed that top cytotoxic effector markers, such as NKG7 and CCL5, are highly present in the most abundant clone and also linked with neutropenia. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Case study III: B-cell antibody synthesis
Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
- PBMC samples from small-cell lung cancer (SCLC) patients were analyzed using Cellecta’s single-cell AIR-BCR-Mark30 profiling assay.
- Tumor-specific B-cells were isolated from antigen-enriched B-cells via peptide tetramers and single-cell FACS sorting.
- A scAIR assay for BCR (IGH, IGK, and IGL chains) was run to obtain B-cell immunoglobulin variable binding domain sequences along with crucial B-cell marker genes.
- Target BCR full-length sequences were used to express in a human IgG backbone to synthesize tumor-specific antibodies.
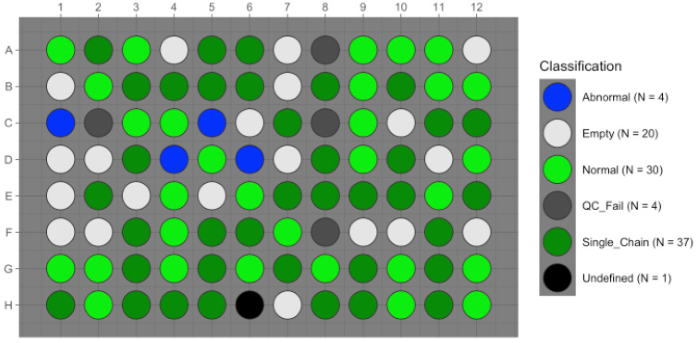
Fig 6. ScAIR analysis shows the IGH-IGL chain pairs used to engineer tumor-specific antibodies from ‘Normal’ wells. Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
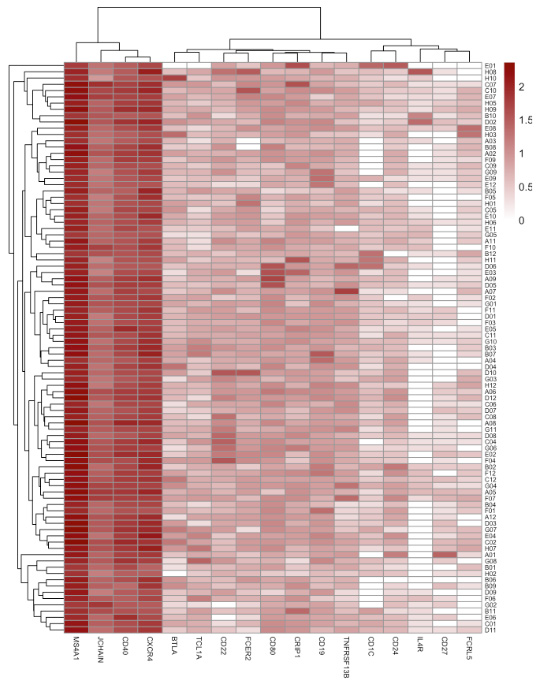
Fig 7. Integrated gene expression analysis of antigen-enriched B-cells from an SCLC patient reveals key B-cell immune markers (JCHAIN, MS4A1, CD40, and CXCR4). Image Credit: Image courtesy of Alex Chenchik et al., in partnership with ELRIG (UK) Ltd.
Discussion
Sensitive, single-cell detection
Cellecta has optimized the multiplex RT-PCR protocol to provide sensitivity high enough to detect and profile T-cell receptor transcripts from sorted or serially diluted T- or B-cells. Low crossover between wells and VBC-based uniform clonotype detection enables the accurate identification of sorted T/B cells, such as antigen-activated cells labeled with specific MHC-peptide antigens.
Chain pairs & immunophenotype
The first-of-its-kind, 96-well plate-based single-cell AIR profiling assay combines TCR/BCR chain full sequence with paired-chain information and the phenotype of cells (e.g., cell subtype) in a single multiplex assay, eliminating the need for specialized instruments such as microwell arrays or droplet microfluidics-based single-cell platforms. Stand-alone TCR and BCR chain-pairing assays are also available for human, mouse, or hybrid human-mouse.
Cost-effective, efficient strategy
For researchers working with hundreds to thousands of rare cells, or have activated cells for which they need chain-pairing -- with our without immunophenotyping information -- this method would be ideal due to its cost-effectiveness (~10 cents/cell) and easy-to-use library prep workflow that allows sample pooling of 96-wells into a single reaction tube, thereby allowing one to scale the number of plates easily run in one day.
Useful for epitope discovery and antibody synthesis
Demonstrated application in identifying paired single-cell receptor chains for TCR-based epitope discovery, and for reconstructing full-length BCR sequences to enable the synthesis of tumor-specific antibodies.
About Cellecta
Cellecta, Inc. was founded in April 2006 by one of the chief scientists from Clontech Laboratories. The company was borne out of the need for higher quality, more advanced shRNA and other lentiviral libraries. Our goal was to develop advanced high-throughput (HT) genetic screen technologies and their applications for the discovery and functional characterization of novel therapeutic targets and drugs.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 26, 2025