This article and associated images are based on a poster originally authored by Sabrina Heaton and presented at ELRIG Drug Discovery 2025 in affiliation with AstraZeneca.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
- Protein mislocalization accounts for ~16 % of all disease-causing missense variants.1
- Assays to detect protein location within the cell are generally tailored to a specific target or require significant resources and expertise to implement.
- Split-receptor systems, where two parts of a receptor must interact to generate a signal, represent an adaptable alternative approach.2
- Unlike most split-receptor systems, the FAST system detects interaction of one primary partner with different secondary partners, wherein each primary-secondary pair results in a different FAST protein that can then bind to a specific fluorogenic dye.3
- For example, a CFAST-tagged target interacting with a ‘green’ NFAST partner under one condition and a ‘red’ NFAST partner under another, results in either a greenFAST protein that binds to ‘Lime’ dye or a redFAST protein that binds to ‘Coral’ dye.3,4
- Adaptation so that a CFAST-tagged target pairs with a different NFAST partner in each subcellular location would result in location-specific FAST proteins that can be identified by their dye-binding preferences.
Methods
- Tested recombinant production of complete FAST proteins (redFAST and greenFAST) and their truncated ‘NFAST’ variants as controls.
- Characterized the sensitivity and specificity of three dyes (Lime (green), Coral (red), and Poppy (far-red)) in binding to recombinant redFAST and greenFAST proteins to identify unique excitation/ emission wavelength pairs for the detection of dye binding.
- Added CFAST motif to known peroxisomal, mitochondrial, and cytosolic control proteins and red, green, or far-red NFAST domains to a scaffold protein (MBP) containing a peroxisomal, mitochondrial, or cytosolic targeting sequence at the N-terminus.
- Introduced CFAST and NFAST pairs by transient transfection of HEK cells, and subsequently demonstrated formation of complete FAST proteins, as detected by binding of Lime, Coral, or Poppy dyes.
- Designed alternative plasmids for stable expression of NFAST domains in their respective subcellular locations to generate a stable cell line for the detection of relative amounts of a CFAST-tagged target in each subcellular location.
Results

Figure 1. Proposed FAST-based assay to detect subcellular localization of target protein. A. Stable cell line containing three different NFAST variants – NFASTred directed to the mitochondria, NFASTgreen directed to the peroxisome, and NFASTfar-red remaining in the cytosol. Without the addition of the CFAST peptide, the three NFAST variants do not bind any of the FAST dyes (Coral, Lime, Poppy), and so no fluorescence signal is generated. When the target protein is introduced through transient transfection, its CFAST peptide tag is able to complement the NFAST variants – generating FAST protein that either: binds to Coral when it pairs with NFASTred in the mitochondria (B); binds to Lime when it pairs with NFASTgreen in the peroxisome (C); and/or binds to Poppy when it pairs with NFASTfar-red in the cytosol (D). Image Credit: Image courtesy of Sabrina Heaton et al., in partnership with ELRIG (UK) Ltd.
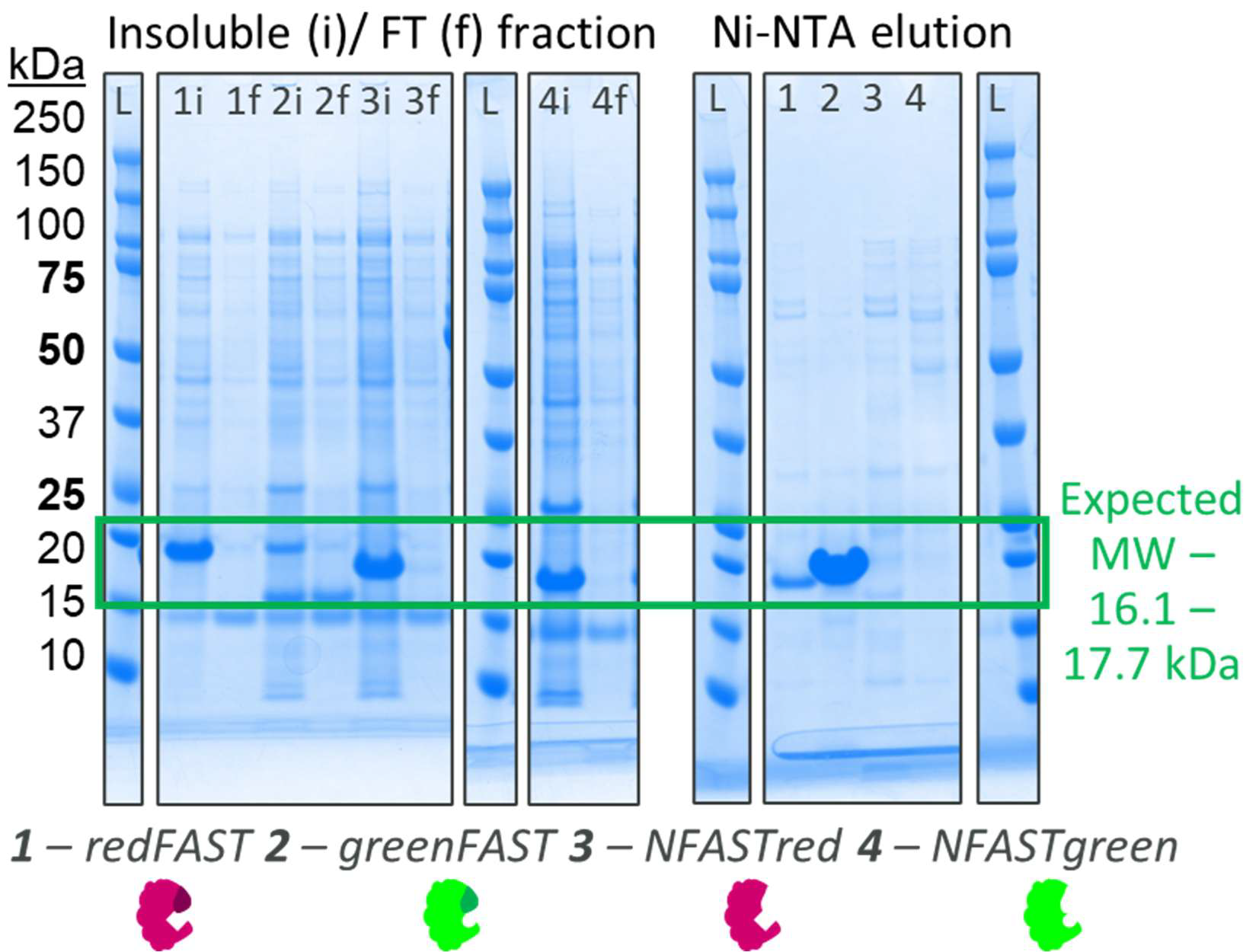
Figure 2. Expression screen of FAST proteins in E. coli. Complete FAST proteins (redFAST (1) and greenFAST (2)) are soluble and highly expressed in E. coli, but removal of the ‘CFAST’ sequence from them, to form either NFASTred (3) or NFASTgreen (4), results in insoluble recombinant protein despite high levels of expression. Image Credit: Image courtesy of Sabrina Heaton et al., in partnership with ELRIG (UK) Ltd.

Figure 3. Comparison of fluorescence signal generated by greenFAST and redFAST binding to Lime, Coral, and Poppy dyes. Coral dye binds to greenFAST and redFAST, with varying signal strength at different wavelengths (left). Lime dye binds much more strongly to greenFAST than redFAST across a broad range of wavelengths (centre). The signal from Lime is 10x the signal from Coral at the same dye and protein concentrations (1 μM each). The binding of Poppy dye is low with greenFAST and not detected with redFAST (right). All three dyes show minimal fluorescence in the absence of FAST proteins (blue). Image Credit: Image courtesy of Sabrina Heaton et al., in partnership with ELRIG (UK) Ltd.

Figure 4. Detection of FAST proteins in HEK cells transfected with NFAST and CFAST control pairs. Perox. = peroxisomal. Mito. = mitochondrial. Cytos = cytosolic. The highest signal occurs when NFAST and CFAST are targeted to the same subcellular location. Image Credit: Image courtesy of Sabrina Heaton et al., in partnership with ELRIG (UK) Ltd.
Conclusion
Pairing of location-specific NFAST variants with a CFAST-containing target protein allows differentiation of the target protein's subcellular localization. After confirmation of NFAST and CFAST expression in the relevant subcellular fractions using Western Blot, the next stage of this project involves generating an intact, stable cell line containing the three NFAST proteins and then applying the validated assay to quantify organelle distribution of a model protein, where mislocalization of disease variants is well-established.
Once validated as a robust assay for detecting protein subcellular localization, the FAST system holds significant promise as a screening platform for identifying and correcting disease variants that cause protein mislocalization.
References
- Lacoste, J., et al. (2024). Pervasive mislocalization of pathogenic coding variants underlying human disorders. Cell. DOI: 10.1016/j.cell.2024.09.003. https://linkinghub.elsevier.com/retrieve/pii/S0092867424010213.
- Bae, J., et al. (2024). Split Proteins and Reassembly Modules for Biological Applications. ChemBioChem, 25(10). DOI: 10.1002/cbic.202400123. https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cbic.202400123.
- Tebo, A.G., et al. (2020). Orthogonal fluorescent chemogenetic reporters for multicolor imaging. Nature Chemical Biology. DOI: 10.1038/s41589-020-0611-0. https://www.nature.com/articles/s41589-020-0611-0.
- The Twinkle Factory. (2025). The Twinkle Factory. (online) Available at: https://www.the-twinkle-factory.com/.
Acknowledgements
The FAST system (proteins and dyes) was developed by Prof. A. Gautier and colleagues (now marketed through The Twinkle Factory), and this work reflects the use of the commercially available products – AstraZeneca was not involved in the discovery, design, or development of these products.
About AstraZeneca
AstraZeneca is a global, science-led patient-focused pharmaceutical company that focuses on transforming the future of healthcare by unlocking the power of what science can do for people, society and the planet.
Science defines who we are. We push the boundaries of science to deliver life-changing medicines. It is why we come to work every day and is part of our DNA. We are committed to operating sustainably, in a way that recognises the interconnection between business growth, the needs of society and the limitations of our planet. We want the way we work to be inclusive, open and collaborative. This approach runs through all that we do.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics, and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free of charge to attend!
Our values
Our values are to ensure the highest quality of content, make it readily accessible to all, and maintain an inclusive organization that serves a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate, and collaborate on an open-access basis. We achieve this through the provision of world-class conferences, networking events, webinars, and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.
Last Updated: Dec 12, 2025