By Shelley Farrar, MSc, BSc
Full genomes of several organisms have been sequenced in the past fifteen years, including the human genome in 2004. These studies were completed using Sanger DNA sequencing, which has a limited throughput and high cost meaning the human genome took fifteen years to sequence and cost nearly three billion dollars. Such limitations have meant that there has been a recent trend towards developing new high-throughput DNA sequencing techniques that allow DNA to be sequenced quickly and cheaply.
There are various high-throughput DNA sequencing techniques but they fundamentally involve:
- template preparation (through isolating and purifying the original DNA fragments and then creating a DNA library)
- clonal amplification (forming multiple copies by loading the library onto a flow cell then amplifying the fragments into clusters)
- parallel sequencing (simultaneously sequencing DNA templates without the requirement for physical separation)
Roche/454 pyrosequencing
One such application is via the Roche/454 pyrosequencer, which is one of the earliest high throughput DNA sequencing techniques. The method utilizes pyrosequencing, which allows for sequencing-by-synthesis as the sequence read out can be achieved at the same time as the sequence is extended. Therefore electrophoresis, as used in Sanger sequencing, is not needed to generate a nucleotide read out of the output.
During pyrosequencing, one nucleotide at a time is washed over copies of the sequence being determined, with the complimentary nucleotides being incorporated onto the template strand. The nucleotide additions release a light signal that can then detect the location and sequence of the nucleotide being incorporated.
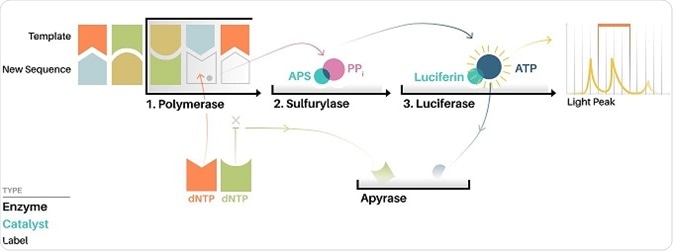
Image: How Pyrosequencing works illustration. ©Jacopo Pompilii, DensityDesign Research Lab. License: Creative Commons Attribution-Share Alike 4.0 Internationa
While this type of sequencing is faster and cheaper than Sanger sequencing, there is a known issue of homopolymer errors where there is a difficulty in distinguishing a run of bases in a sequence that are identical, such as the sequence GGGG (i.e. the guanine quartet).
Illumina Genome Analyzer
The Illumina Genome Analyzer also has a sequencing-by-synthesis concept where the reaction is stopped after each base, a fluorescent dye is used to read the base label and the sequence reaction is then continued with the next base.
During clonal amplification the new strand is covalently bound to the flow cell. This new strand can bend and attach to an oligonucleotide that is complementary to the adaptor sequence at the free end of the new strand. A second covalently bound reverse strand can then be synthesized, which is called a bridge amplification, and can be repeated to form clusters.
More than 200 million clusters per run can be formed and 150 nucleotides can be sequenced from both ends of a fragment. This is accomplished by washing away the synthesized sequence, repeating the bridge amplification cycle for the reverse of the strand, removing the starting strand and adding a new sequencing primer for the second read. This allows for twice the amount of sequenced data to be generated.
However, there is a high background error rate because the production of the library and flow cell requires in vitro amplification steps. The method may also incorrectly establish lint, dust and chemical particles as clusters, though the low sequence complexity that is resulted can be easily identified.
Overview of Illumina Sequencing by Synthesis Workflow | Standard SBS chemistry
Ion torrent and Ion proton sequencing
Another technique is Ion Torrent and Ion proton sequencing, which unlike Roche/453 and Illumina techniques does not use optical signals during sequencing. Hydrogen (H+) ions are detected instead. When a deoxynucleotide is added to a DNA polymer a H+ ion is released. This can be detected through a decrease in pH and changes in the pH can be used to determine the base added, and so the sequence can be read. This technique also suffers from the homopolymer error problem like the Roche technique as sections where the same base is repeated are difficult to define.
Applications of high-throughput DNA sequencing techniques
In 2014, a new generation of Illumina Genome Analyzer was created that can efficiently sequence 45 human genomes a day for 1000 US dollars each. This means that genome sequencing for medical and personal applications is closer to affordability.
By the application of high-throughput DNA sequencing techniques, it is possible to identify the variants that cause genetic disorders. As the technologies become cheaper and more efficient, their application will become increasingly common and a new age of personalized (or precision) medicine will be created.
Sources:
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3831009/
- www.ebi.ac.uk/.../what-next-generation-dna-
- https://www.ncbi.nlm.nih.gov/pubmed/20486139
- http://www.cell.com/molecular-cell/abstract/S1097-2765(15)00340-8
Further Reading
Last Updated: Feb 26, 2019