This article and associated images are based on a poster originally authored by Poonam Shah, Mat Calder, Emma Stanway, Ben Durham, Aishwarya Sundaresh, Ásta-Björk Jonsdottir, Amy Prosser, Miguel Coelho, Bethany Atkinson, Aimee Blair, Jack Cobb, Marta Falcicchio, Ruben Alvarez Fernandez, Emma Ford, Alberto Moreno de la Gandara, Ilaria Giovannelli, Penny Hayward, Sajaana Jeyaseelan, Matt Jones, Navrohit Kandola, Mick Knaggs, Tabitha Morgan, Kathleen Santos, Chloe Tarry, Eleanor Thompson, Lucy Walker, Robert Yan, Willie Yen, Benedict Cross, Rich Boyce and Christian Dillon and presented at ELRIG Drug Discovery 2025 in affiliation with PhoreMost Limited.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
Targeted protein degradation (TPD) modalities hijack the cell protein recycling pathways to reduce levels of disease associated proteins, providing further potential therapeutic benefits over traditional small molecule inhibitors.
Molecular glues or heterobifunctional molecules can induce proximity between an E3 Ligase and a Protein of Interest (POI) and lead to its ubiquitination and degradation using the ubiquitin proteasome system (UPS).
The human genome encodes over 600 ligases, yet the majority of degraders in clinical and preclinical development rely predominantly on the recruitment of a single E3 ligase, Cereblon. Arising resistance, toxicities, and restricted Protein-of-Interest scope may limit clinical potential, highlighting the need to uncover novel TPD mechanisms.
Through the PhoreMost SITESEEKER® screening technology, we have uncovered several novel E3 ligases for the development of next-generation degraders to move beyond Cereblon. The PhoreMost drug discovery pipeline has progressed multiple small molecule binders that have been incorporated as warheads in heterobifunctional molecules to effectively degrade selected POIs.
We demonstrate that Ligase X, which functions through the CUL1/SKP1 SCF complex, can be hijacked to degrade various oncology targets, such as BRD4, both in cells and in vivo.
We found we can also hijack members of the CUL2 complex, with successful degradation of the inflammation target TYK2 using our KLHDC2 binder. We also describe high-affinity small molecule binders against an N-degron-based Ligase Y.
Discovery of these novel E3 ligases has the potential to expand the development of a wider range of therapeutic targets.
PHOREMOST E3 ligase chemical platform
- The PhoreMost drug discovery pipeline identified potent small molecule binders to multiple E3 ligases that have been used as warheads in heterobifunctional molecules
- Novel E3 ligase-based heterobifunctional molecules expand the range of proteins-of-Interest that can be degraded for use in TPD modalities across multiple therapeutic areas including oncology and inflammation
- Ligase X is overexpressed in a large subset of tumors supporting cancer selective degradation of a range of proteins-of-interest
- KLHDC2 is widely expressed in a range of tissues and known to degrade a variety of natural substrates, suggesting potential for a broad scope of proteins-of-interest to target
- Ligase Y is an N-degron pathway ligase that has a ubiquitous expression profile. N-Degron ligases are an underutilized modality in TPD enabling unprecedented proteins-of-interest degradation
Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
Results
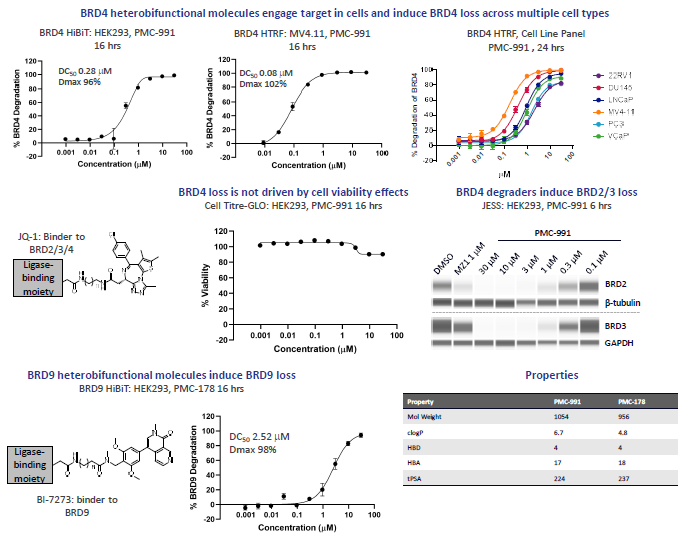
Figure 1A. Ligase X-based heterobifunctional molecules can effectively degrade a range of proteins of interest. BRD4 and BRD9 heterobifunctional molecules were synthesised exploiting two independent exit vectors and a range of linker lengths/types, with the proteins-of-interest binders indicated. Optimised degraders engaged with the target in cells, leading to ubiquitination (not shown) and rapid degradation in multiple cell models. Effective degradation of a diverse range of proteins of interest was demonstrated. Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
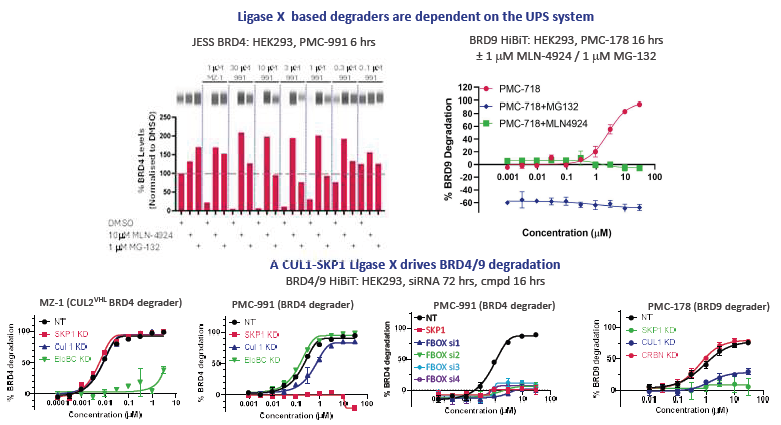
Figure 1B. Ligase X functions through a differentiated TPD mechanism-of-action involving the CUL1-SKP1 SCF complex. BRD4/9 degradation is dependent on the Cullin-UPS pathway (MLN-4924/MG-132 rescues degradation). siRNA experiments demonstrated a key functional dependency on the CUL1-SKP1 SCF complex. Ligase X is highly upregulated in many cancers (not shown), raising the possibility of enhanced cancer-selective degradation. Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
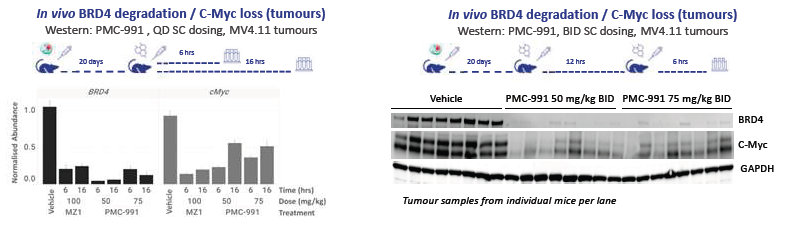
Figure 1C. Ligase X degraders show effective in vivo degradation of BRD4 in mouse xenografts. Mice were dosed subcutaneously with tissue or MV4.11 xenografted tumors harvested as indicated. Significant reductions in BRD4 levels in BRD4/C-Myc levels in implanted tumors were observed. BID dosing leads to complete loss of tumor-associated BRD4 levels. Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
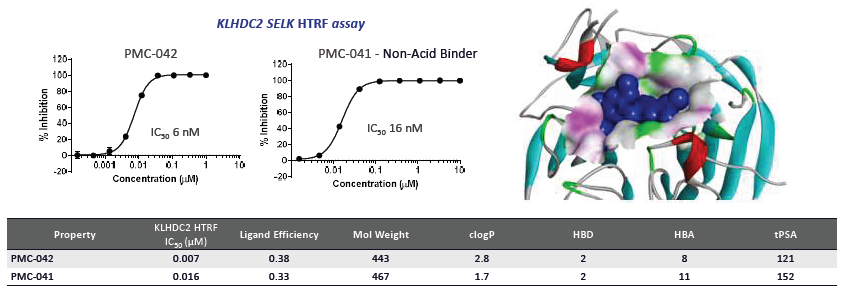
Figure 2A. Optimisation of high affinity small molecule binders to KLHDC2. A virtual screen of ~6M compounds and subsequent validation/optimisation led to three series of high-affinity small molecule binders (e.g., PMC-042). Biochemical Assays (HTRF, SPR, and FP) were used to measure binding affinity to KLHDC2. Soaked crystal structure of KLHDC2 bound to PMC-908 at a resolution of 2.5 Å. Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
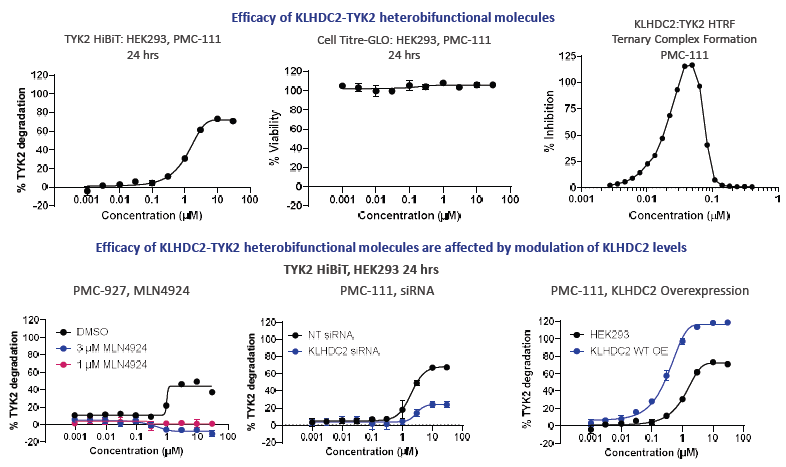
Figure 2B. KLHDC2 heterobifunctional molecules induce ternary complex formation to drive loss of TYK2. KLHDC2 based degraders can lead to a loss of 80 % TYK2 levels, without any loss in cell viability. Ternary complex formation of TYK2 and KLHDC2 can be detected by HTRF. The degradation of TYK2 is proteasomal dependent and can be rescued with a knockdown of KLHDC2. Overexpression of KLHDC2 can boost potency of the degraders and can lead to 100 % loss in TYK2 levels. Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
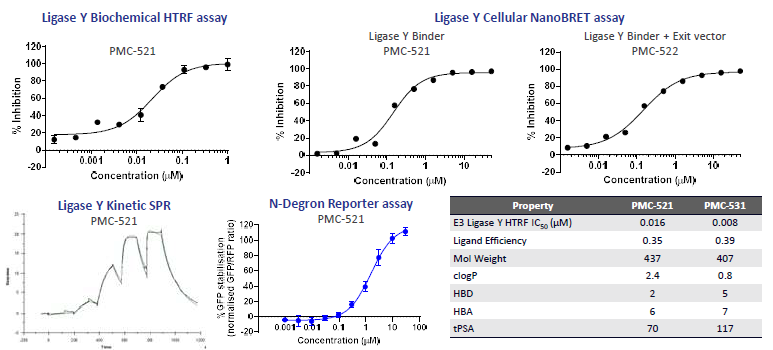
Figure 3A. Optimisation of high affinity small molecule binders to N-degron based Ligase Y. A DEL screen of ~140B compounds and subsequent validation/optimization led to 2 series of high affinity small molecule binders (e.g. PMC-521). Biochemical (SPR and HTRF) and cellular (NanoBRET) assays were used to measure binding affinity to Ligase Y. Binding affinity to Ligase Y is maintained upon addition of exit vector to binder. N-degron assay was used as an orthogonal assay to validate binding to Ligase Y. Image Credit: Image courtesy of Poonam Shah et al., in partnership with ELRIG (UK) Ltd.
Conclusion
- SITESEEKER® platform identifies novel E3 ligases for use in TPD, providing a differentiated MOA to VHL/Cereblon-based degraders.
- Heterobifunctional molecules can degrade a diverse set of proteins of interest using Ligase X, which operates through the CUL1-SKP1 SCF complex.
- Ligase X based degraders are highly active in tumor-bearing mouse models.
- KLHDC2 based degraders can be used to degrade a variety of proteins-of-interest including oncology target BRD4 and inflammation target TYK2.
- Highly potent small molecules that bind to N-degron-based Ligase Y have been designed and optimised.
About PhoreMost

PhoreMost is a new-model drug discovery company based in Cambridge, UK: Using its core expertise to open up new ‘druggable’ target space and working with a global network of co-invested academic and industrial collaboration partners, we aim to bring a wide array of novel ‘targeted’ therapies more efficiently to market and pass these cost savings onto patients.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free of charge to attend!
Our values
Our values are to always ensure the highest quality of content, that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate, and collaborate on an open-access basis. We achieve this through the provision of world-class conferences, networking events, webinars, and digital content.
Sponsored Content Policy: News-Medical.Net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.
Last Updated: Dec 1, 2025