Nanoparticle tracking analysis (NTA) is a method for visualizing and analyzing particles in liquids. NTA suspends small particles (~10-1000 nm) in a liquid, this allows for the determination of a size distribution profile.
Cobalt nanoparticles covered by aluminum with a scale bar. Image Credit: Georgy Shafeev / Shutterstock
Performing Nanoparticle Tracking Analysis
The particles that are to be analyzed are placed into a sample chamber. A laser beam is passed through the chamber and the suspended particles in the laser’s path scatter the light, which allows them to be visualized. A camera is used to take a video file of the particles moving under Brownian motion. This is very cost and time effective as it allows simultaneous measurement of multiple characteristics of the particles.
The rate of particle movement is related to a sphere equivalent hydrodynamic radius which is calculated through the Stokes-Einstein equation. The size of each particle is calculated individually, which provides detail regarding the whole population of particles. The recording provided can be analysed with a range of different outputs.
NTA is currently used for particles from 10 to 1,000 nm in diameter. Analysis of particles as small as 10 nm is only possible for particles that are made from material with a high refractive index. Analysis of particles bigger than 1,000 nm is not possible due to the Brownian motion of large particles (Larger particles move slowly which reduces the accuracy). The solvent viscosity can also determine the upper size limit for particles during an NTA experiment because it influences the movement of the particles.
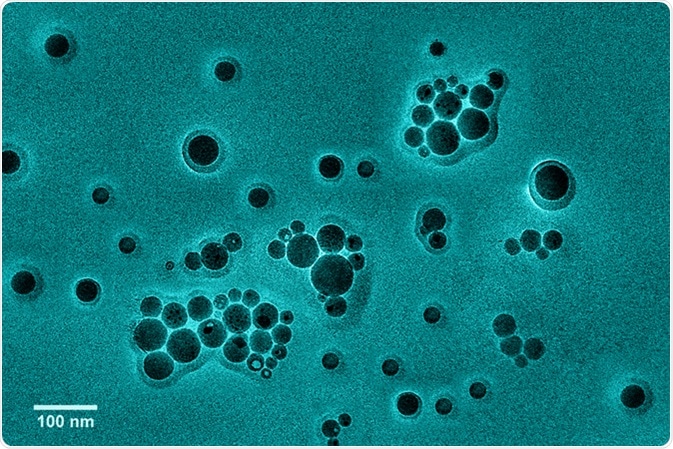
Vesicle size and concentration by Nanoparticle Tracking Analysis
Uses of Nanoparticle Tracking Analysis
Applications of NTA include:
Protein Aggregation Studies
Protein aggregation can occur during many biological manufacturing processes including Cell cultures, purification, and formulation. NTA can quickly and accurately assess the amount of protein aggregation that has occurred.
Nanotoxicology
Information provided by NTA can be used to assess the potential toxicity of a drug. The particle size, concentration, and aggregation influence how it interacts within the body and can lead to health implications.
Viral vaccine research and development
Knowing the particle size distribution of viruses and bacteriophages is critical for the research and development of vaccines. An NTA experiment can provide the total viral count, which allows gives a better understanding of infectious viruses as well as non-infectious viruses.
Development of drug delivery systems
Designing and manufacturing targeted drug delivery systems benefits from knowing about the distribution of nanoparticle size. New and innovative vaccines can be made from polymeric nanoparticles, exosomes, and liposomes with the help of NTA instruments. The size and distribution of nanoparticles in the medicines can affect skin diffusion, immune response, the release of vaccine components, and call uptake of drug delivery within the immune system.
Extracellular vesicle analysis
Characterization of extracellular vesicles can be determined by NTA. Extracellular vesicles have the potential to be used for the diagnostics and treatment of different diseases. However, the heterogeneity of extracellular vesicles and the choice of NTA instrument settings may cause variation in the analysis results, which limits their biomedical use.
Conclusion
In conclusion, NTA has many uses in the field of life sciences. The applications it has can aid with the research into the disease, development of drug treatments, and assessing the toxicity of certain compounds. NTA has clear advantageous and disadvantageous that can be exploited to provide better and more accurate results. Continued research into NTA will allow for more discoveries using it and for more methods of acquiring more accurate results.
Further Reading
Last Updated: Aug 23, 2018