A global analysis reveals that bacteria in food animals and humans are sharing resistance genes through the use of agricultural antibiotics, highlighting an urgent need to rethink farm drug regulations.
 Study: The ionophore resistance genes narA and narB are geographically widespread and linked to resistance to medically important antibiotics. Image Credit: Thammachak Sotiya / Shutterstock
Study: The ionophore resistance genes narA and narB are geographically widespread and linked to resistance to medically important antibiotics. Image Credit: Thammachak Sotiya / Shutterstock
In a recent article published in mSphere, researchers examined the global distribution of ionophore resistance conferred by two genes, narA and narB, and their genetic linkages to genes associated with antimicrobial resistance (AMR). They found that 2,442 bacterial isolates from 51 countries contained narAB, often in conjunction with genes that confer resistance to critical human antibiotics. These results highlight the significant risks of co-selection for human-relevant AMR through the use of ionophores in agriculture.
Background
Ionophores are antibiotics commonly used in animal agriculture, primarily as anti-coccidials in poultry and for growth promotion in cattle and swine. In 2022, they accounted for 37% of antibiotics used in U.S. food-producing animals.
Because ionophores are not used in human medicine due to toxicity, they are classified as low-risk for contributing to human AMR and are less stringently regulated than other antibiotics.
However, recent studies have raised concerns that ionophores could indirectly promote AMR through co-selection. This can occur in two ways: through physical linkage, where the genes are located near each other on the same piece of DNA (like a plasmid), or when they are present in the same genome of a largely clonal bacterial species. One focus has been on the narAB operon in Enterococcus faecium, which provides resistance to several ionophores and has been found on plasmids carrying vancomycin, erythromycin, and tetracycline resistance genes.
Co-selection could occur if such genes are transferred together, even in the absence of direct antibiotic pressure from MIAs. For example, a decline in vancomycin-resistant Enterococcus was observed in Norway after the use of ionophores in poultry ceased. Despite these signals, the full geographic spread and the consistency of AMR associations of narAB have remained largely unquantified.
About the study
To address this knowledge gap, researchers used a publicly available global dataset to assess how widespread narAB is and to what extent it is genetically linked to clinically relevant AMR genes. The search focused on annotated bacterial genomes containing narA, and only isolates also containing narB were retained, resulting in 2,442 qualifying genomes. These were analyzed for host organism (such as human, poultry, or cattle), bacterial species, associated AMR genes, and country of origin.
To assess the potential for co-selection, the team measured the frequency with which narAB-positive genomes also contained resistance genes or mutations associated with MIAs. They calculated the average number of resistance genes and mutations per genome. They used statistical association tests to determine if particular AMR determinants were more likely to occur in narAB-positive isolates of Enterococcus faecalis and E. faecium. Genes and point mutations with strong associations were flagged as potential evidence of co-selection.
The researchers also visualized the geographic spread and host species using maps and bubble plots, summarizing the prevalence of narAB in different bacterial species and host organisms, including humans. Their analysis focused on presence/absence trends, given uneven global sampling, and did not attempt to compare prevalence across countries.
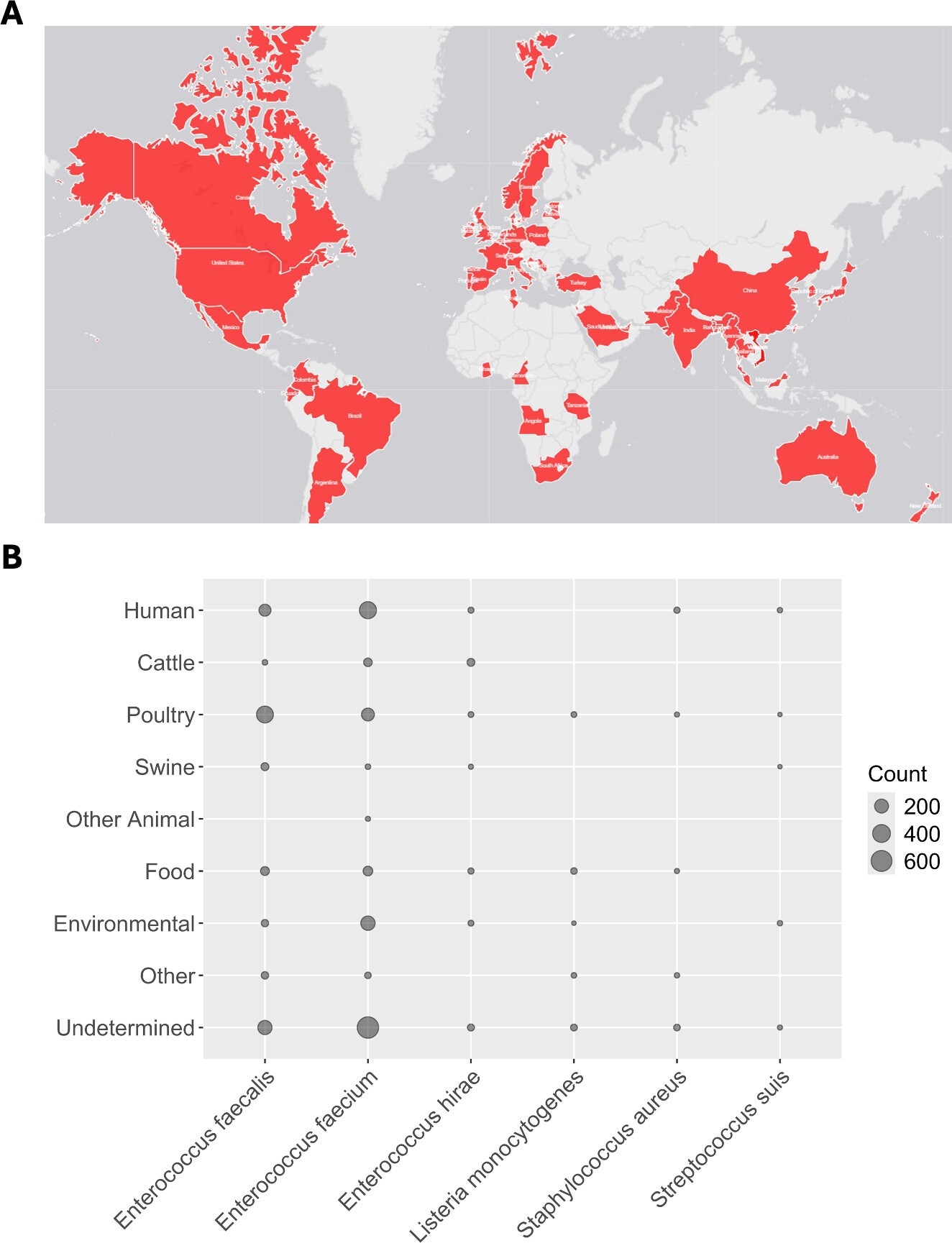
Distribution of narA/B. (A) World map showing the geographic distribution of narAB. (B) Distribution of narAB amongst bacterial and host species. Bubble size is proportional to the number of detected isolates. Four bacterial species are excluded due to total sample sizes of less than 5 (Campylobacter jejuni, Clostridium perfringens, Listeria innocua, and Streptococcus agalactiae).
Key Findings
The study found narAB genes in 2,442 bacterial isolates from 51 countries, demonstrating global presence across all continents except Antarctica. The genes were identified in 10 bacterial species, predominantly Enterococcus faecalis and E. faecium.
Isolates came from diverse hosts, including humans, cattle, swine, and poultry, and over 500 human-derived isolates carried narAB, challenging the assumption that these genes are limited to non-human contexts and underscoring the interconnectedness of human and animal health.
Genomic analysis revealed that narAB-positive isolates contained an average of 8.26 additional AMR genes and 2.13 point mutations conferring resistance. Common AMR genes included ermB (erythromycin resistance), tet(M) (tetracycline resistance), and aac(6′)-I (aminoglycoside resistance). Mutations associated with resistance to drugs like daptomycin and ampicillin were also detected.
In E. faecalis, 13 AMR genes, including all vanA cluster genes (linked to vancomycin resistance), showed strong positive associations with narAB. In E. faecium, narAB was associated with 11 AMR genes and 4 resistance-linked mutations.
These findings reveal a strong statistical pattern suggestive of co-selection, where ionophore resistance genes are genetically linked to AMR genes relevant to human health. The evidence supports the idea that ionophore use in agriculture may unintentionally select for broader AMR, including resistance to critically important antibiotics.
Conclusions
This study challenges the long-standing assumption that the use of ionophores in livestock is unrelated to human antimicrobial resistance risks. The widespread detection of narAB in multiple countries, bacterial species, and even human-derived isolates suggests that ionophore resistance is not confined to animal agriculture.
The consistent genetic associations between narAB and resistance to medically important antibiotics, especially in Enterococcus faecalis and E. faecium, highlight how genetic linkage provides a clear pathway for co-selection, where ionophore use may indirectly promote clinically relevant AMR.
A key strength of the study is its global scope, using a large publicly available dataset to provide comprehensive genomic and geographic insights. However, uneven sampling limits the comparative assessment of prevalence across countries. Additionally, although the study reveals associations between narAB and AMR genes, it cannot definitively establish causality or the primary directionality of gene transfer; however, transfer from farm animals to humans appears most likely.
Overall, the findings suggest ionophore use may pose an underrecognized AMR risk, emphasizing the need to reconsider how such antibiotics are regulated and highlighting that even drugs not used in human medicine can contribute to the broader AMR crisis.
Journal reference:
- The ionophore resistance genes narA and narB are geographically widespread and linked to resistance to medically important antibiotics. Ibrahim, A., Au, J., Wong, A. mSphere (2025). DOI: 10.1128/msphere.00243-25 https://journals.asm.org/doi/10.1128/msphere.00243-25