Combining image analysis with a three-dimensional microscopy technique allows researchers to quantify new or little-understood cell biology phenomena, according to a new study publishing December 19 in the open-access journal PLOS Biology by Mathieu Frechin of the Swiss company Nanolive and colleagues. The technique is likely to help explain little-understood aspects of the behavior of cell organelles – the functional compartments within a cell.
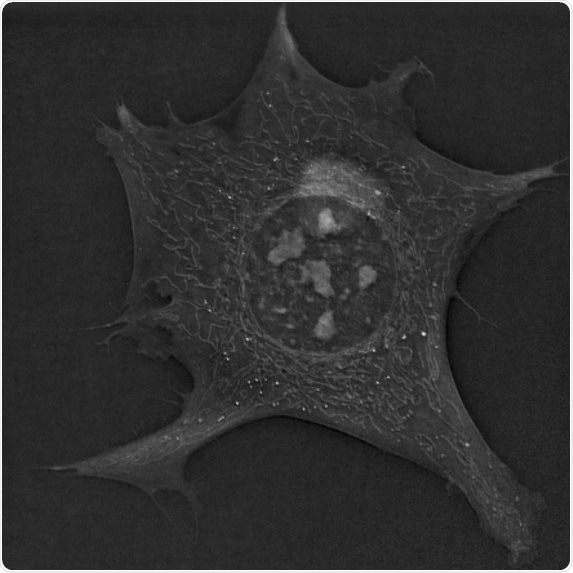
This image displays a single 3T3-L1 pre-adipocyte cell imaged with holotomography. The remarkable feature of this image is the beautiful mitochondrial network, elongated and well connected. Image Credit: Hugo Moreno and Dr. Mathieu Frechin
A wide variety of techniques is available to study living cells, but many of them have intrinsic limitations, such as the need for damaging levels of light, or the use of interfering dyes, or poor contrast and resolution.
A relatively new form of microscopy, called holo-tomographic microscopy, surmounts some of these limitations, by splitting a light beam, diverting one part and letting the other pass through the sample. When the two parts are recombined, differences in their wave forms can be used to create an image based on differences in the refractive index of the cell’s components. By tipping the sample, multiple such images can be combined to create a three-dimensional picture of the cell. The whole process is done without dyes and at very low levels of light, preserving the structural and behavioral integrity of the cell during image capture.
In the current study, the authors applied multiple image-processing techniques to extract quantitative data from their holo-tomographic images. Applying their techniques, they quantified for the first time the flux of fat droplets within a whole cell, measuring their rate of formation and changes in mass over time, and revealing new aspects of their dynamics, including the synchronization of swelling among a subset of droplets.
The ability of the microscopic technique to capture data from the entire cell simultaneously allowed the authors to provide important new information about the intriguing but little-understood process of organelle spinning. Here, in the run-up to mitosis (the stage of the cell cycle in which the nucleus divides in two), the researchers show that the entire nucleus rotates between 80 and 700 degrees over a period of minutes to hours.
The ability to not only observe, but to measure dynamics of undisturbed living cells, through the application of quantification techniques to holo-tomographic microscopy, is likely to lead to discovery of fundamentally new cell behaviors, and new understanding of previously studied phenomena as well."
Mathieu Frechin, Swiss company Nanolive
Source:
Journal reference:
Sandoz, P.A., et al. (2019) Image-based analysis of living mammalian cells using label-free 3D refractive index maps reveals new organelle dynamics and dry mass flux. PLOS Biology. doi.org/10.1371/journal.pbio.3000553.