Researchers in the United States have identified both antigen-specific and antigen non-specific predictive signatures of antibody responses to Moderna’s two-dose coronavirus disease 2019 (COVID-19) vaccine that protects against infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The team found that after both vaccine doses, robust and coordinated immunoglobulin A (IgA) and IgG responses were preceded by bursts of plasmablasts that are specific to the SARS-CoV-2 spike protein, although these bursts occurred earlier and more intensely after the second dose.
The viral spike protein is the main structure SARS-CoV-2 uses to infect host cells and the primary target of antibodies following vaccination or natural infection.
The team from the National Institutes of Health in Bethesda and the Institute for Bioscience and Biotechnology Research in Rockville found that distinct antigen-specific memory B cell populations with varying kinetics also emerged following vaccination.
Susan Moir and colleagues say that these predictive signatures could serve as early indicators of the serologic efficacy of messenger RNA (mRNA)- based COVID-19 vaccines and help to explain the variability in vaccine efficacy across individuals.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Some individuals have weak responses to the current vaccines
The mRNA-based vaccines developed by Moderna (mRNA-1273) and Pfizer-BioNTech (BNT162b2) encode a stabilized ectodomain of the viral spike protein trimer (S-2P) derived from the Wuhan Hu-1 isolate.
Both vaccines have been shown to be highly effective at eliciting strong B cell and antibody responses.
However, some individuals, such as the elderly and transplant recipients, exhibit weak responses to vaccination, which raises questions about the factors that determine these responses and whether early correlates of immunity can be defined.
“Studies on other vaccines have shown that pre-vaccination signatures and early circulating B cell responses involving plasmablasts (PBs) and activated memory B cells (MBCs) can predict the magnitude and longevity of neutralizing antibodies following vaccination,” writes Moir and colleagues.
The researchers say that while the Pfizer-BioNTech vaccine has been shown to elicit robust PB and MBC responses in blood and lymph nodes, the extent to which these responses differ between individuals and whether they are associated with antibody levels have not yet been assessed.
What did the researchers do?
The team evaluated antibody, plasmablast, and B cell responses among 21 healthy individuals uninfected with SARS-CoV-2 who received two doses of Moderna’s mRNA-1273 vaccine.
Blood samples were tested at baseline (pre-vaccination) and then serially over a period of approximately 60 days after vaccination. Paired serum and cellular assays were performed at each timepoint.
Antibody binding to S-2P and its receptor-binding domain (RBD) was measured using a multiplex platform.
What did the study find?
The team observed strong IgG and IgA responses to S-2P and RBD that started around day 10, following the first vaccine. However, these responses varied by 2 to 3 orders of magnitude across vaccinees 28 days following the second vaccine.
Next, the team used a pair of RBD and spike subunit 1 (S1) tetramers to track spike-specific B cell responses. This revealed that RBD+S1+ and single S1+ PBs and MBCs became detectable at day 10 and day 14, respectively, following the first vaccine.
Clustering analysis of 15 antibodies against B cell markers revealed 30 clusters grouped by eight major B cell populations.
Five populations of conventional MBCs and eight nonconventional MBCs were identified, along with IgG (C9) and IgA (C13) PBs. The PB C9 cluster contained the highest proportion of RBD+S1+ cells, followed by C13 and the nonconventional MBC C5.
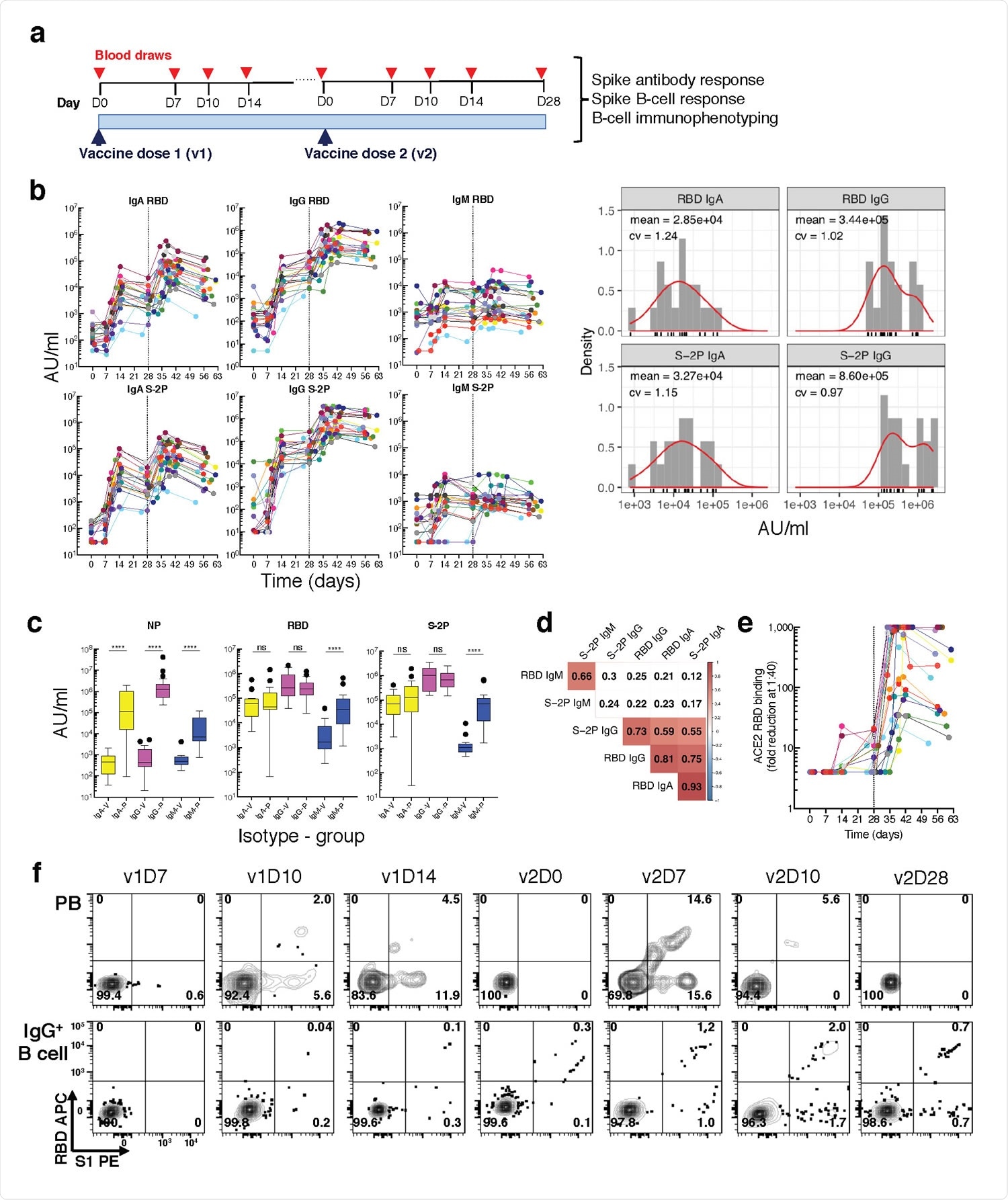
Longitudinal blood sampling and analysis shows robust antibody and early B cell response to mRNA-1273 vaccine. a, Study design with serial blood draws and assays performed at all timepoints on SARS-CoV-2-uninfected vaccinees (n = 21) receiving two doses of the mRNA-1273 vaccine. b, Serum IgG, IgA and IgM binding to S-2P and RBD proteins measured by electrochemiluminescence (ECLIA) longitudinally (left panels), and corresponding histogram and distribution (based on kernel density estimates) at the last timepoint (v2D28) (right panels). c, Peak serum IgG, IgA and IgM binding to S-2P, RBD and N proteins measured by ECLIA in vaccinees (V; n = 21) and COVID503 19 patients (P; n = 21), shown as boxplots. d, Triangular heatmap of correlation between serum antibodies at last measured timepoint (v2D28) in (b). Numbers represent r values. Statistically insignificant correlations (p > 0.05) shown in white. e, Longitudinal inhibition of RBD binding to ACE2 by serum (1:40 dilution) of vaccinees (n = 21). f, Longitudinal binding of S1 and RBD tetramers to PB and IgG+ 507 B cells by flow cytometry shown for a high responder (VAC-611; Extended Data Table 1). Numbers in each quadrant are percentages. Each vaccinee is color509 coded and second vaccine dose indicated by vertical dotted line (b,e). Mann-Whitney test; ****, p < 0.0001 (c). Spearman’s rank correlation (d). AU, arbitrary units; D, day; N, nucleocapsid; ns, not significant; P, patients with severe COVID-19; PB, plasmablasts; RBD, receptor binding domain; S1, spike subunit 1; S-2P, stabilized spike trimer; v, vaccine dose; V, vaccinees.
Longitudinal tracking revealed differences in B cell responses following each vaccine dose
Longitudinal tracking of the vaccinees revealed that RBD+S1+ PBs were first detected at day ten following the first vaccine, with levels of these PBs then subsiding up to five to seven days following the second vaccine.
By contrast, RBD+S1+ MBCs became visible at day 14 following the first vaccine and intensified following the second.
Levels of nonconventional MBC clusters C3 and C6 decreased after each vaccine, while the PB clusters C9 and C13 and the IgA pre-vaccination baseline PB C12 sharply increased.
Spike-specific B cell frequencies showed varying patterns of temporal responses. Two-peak responses were observed for the PB clusters C9 and C13 and the nonconventional MBC clusters C11 and C6, with stronger increases following the second versus first vaccine.
At day 10 following the first vaccine, both C9 and C13 were positively associated with both anti-RBD and anti-S-2P IgA titers, while MBC clusters C3, C11, and C24 were positively associated with anti-S-2P IgG titers as early as day 7.
At day 7 and day 10 following the second vaccine, C9 correlated with anti-RBD IgG and IgA titers, while C13 correlated with anti-S-2P IgA titers at day 7.
These signatures could help predict serological efficacy of mRNA vaccines
“Together, our analyses revealed both antigen non-specific and spike-specific predictive signatures of antibody responses,” writes Moir and colleagues.
“These signatures could help predict and monitor the serological efficacy of SARS-CoV-2 mRNA vaccines and pave the way to a better understanding of weakened responses to this novel vaccine platform, such as those associated with age and chronically compromised immune systems,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Moir S, et al. Pre-vaccination and early B cell signatures predict antibody response to SARS-CoV-2 mRNA vaccine. medRxiv, 2021. doi: https://doi.org/10.1101/2021.07.06.21259528, https://www.medrxiv.org/content/10.1101/2021.07.06.21259528v1
- Peer reviewed and published scientific report.
Kardava, Lela, Nicholas Rachmaninoff, William W. Lau, Clarisa M. Buckner, Krittin Trihemasava, Jana Blazkova, Felipe Lopes de Assis, et al. 2022. “Early Human B Cell Signatures of the Primary Antibody Response to MRNA Vaccination.” Proceedings of the National Academy of Sciences 119 (28). https://doi.org/10.1073/pnas.2204607119. https://www.pnas.org/doi/full/10.1073/pnas.2204607119.