Introduction
An invaluable source of RNA, DNA, and chromatin from historical and clinical samples is formalin fixed paraffin embedded (FFPE) tissue. Globally, tissue banks, hospitals, and laboratories are estimated to store over a billion tissue samples, consisting mainly of FFPE tissue (Tang et al, 2009).
For performing analysis such as immunohistochemistry (IHC), many pathology samples are stored as FFPE blocks. Using RNA and DNA extraction, researchers have already started unlocking the potential of this tissue for epigenetic and genomic analysis. Extensive profiling of genomes is becoming more important, since analyzing larger sample cohorts to investigate many biomarkers used for prognosis and targeted therapies is in line with the current requirements in personalized medicine.
Access to cohorts with related patient information, diagnosis, and treatment outcome is needed to translate the potential of epigenetic profiles for new biomarker discovery and validation. Archived tissue has advantages over frozen or fresh tissue, since it remains viable over a prolonged time period for further analysis as more clinical data and disease outcomes are collected even long after performing biopsies. In order to examine mutations in genes (Beadling et al, 2011, Suz et al, 2011) and gene expression (Fanelli et al, 2011), genotyping technologies have been successfully used on nucleic acids from FFPE tissue.
Due to widespread cross-linking and damage to protein epitopes, as a result of the fixation process and the destruction of these proteins during DNA extraction protocols, utilizing the chromatin from these samples has not been very easy. Conventional DNA extraction protocols can remove proteins (Fan and Gulley, 2001) and frequently involve extraction of phenol in which the protein is separated in the interphase (Pikor et al, 2011).
For high throughput genetic profiling, the constraints are sample quality and availability (Pikor et al, 2011). DNA yield is often low, chemically modified, and highly degraded due to the fixation process and the extensive cross-linking occurring in the preparation of FFPE tissues (Bourgen et al, 2014).
Formalin fixation leads to cross-links between DNA and proteins as well as between the strands of DNA themselves (Lin et al, 2009) which leads to inhibition of PCR and other downstream processes (Gilbert et al, 2007). Extremely low pH and other fixation conditions result in further fragmentation of DNA compounding poor PCR efficiency. Other than the issues with the DNA component of the genetic material, extraction of chromatin from FFPE tissue itself presents some unique challenges.
These challenges are overcome by the Chromatrap® FFPE ChIP kit by employing an optimized buffer system for extraction, resulting in a much higher yield of chromatin, leading to the availability of more protein epitopes to ChIP antibodies. The dual benefits of increased sensitivity and elimination of the need for high chromatin loading in the Chromatrap® ChIP system makes Chromatrap® FFPE ChIP kit the ideal solution for epigenetic research utilizing FFPE tissue.
Method
FFPE sample preparation
In this study, rat uterine tissue (fixed in 10% formalin for 18 hours and soaked in 70% ethanol before embedding in paraffin wax, as shown in Figure 1a) and human breast tissue (Amsbio, Oxford UK), fixed in 10% neutral formalin for 24 hours before embedding in immunohistochemical grade paraffin wax were used as FFPE samples, as shown in Figure 1b.
Figure 1. Rat uterine and human breast tumor FFPE tissue blocks
A microtome (Leica) was used to section each tissue type into 5 µm slices, which were then placed into a microcentrifuge tube. 20 x 5 µm slices of each tissue type were pooled into a microcentrifuge tube per extraction.
Chromatin extraction
The Chromatrap® FFPE ChIP kit protocol was adopted for the extraction of chromatin from the FFPE tissue blocks. By adding 1ml of Paraffin Removal Solution (PRS) to each tube and incubating samples on a rotating platform for five minutes at room temperature (RT), the paraffin wax was removed from each sample. Next, the tubes were centrifuged for five minutes at maximum speed at RT and the supernatant was carefully aspirated. Additional PRS was added to each sample, and the preceding step was performed again to a total of three washes in PRS.
The tissue was rehydrated by addition of 1ml 100% ethanol after aspiration of the final PRS wash, and tubes were subjected to a second incubation on a rotating platform for five minutes at RT. Then, samples were centrifuged for five minutes at full speed at 4 °C followed by careful aspiration of the supernatant. The washing process was again performed with 70% ethanol, 20% ethanol, and finally with sterile distilled water. After aspiration of the distilled water, the pellet was re-suspended in 1ml FFPE Lysis Buffer and incubated for 30 minutes at RT on a rotating platform. The samples were centrifuged for five minutes at maximum speed at 4 °C, the supernatant was aspirated, and the pellet resuspended in 500 µL Digestion Buffer.
The samples were homogenized by sonicating for three cycles of 30s on, 30s off at 42% amplitude and chromatin digested by adding 1µl of Shearing Cocktail. Enzymatic Stop Solution was added after 5min incubation at 37 °C the samples were mixed by pipetting and for five minutes. Centrifugation was performed to collect the pellets, the supernatant was carefully aspirated, and the samples were resuspended in 500 µL FFPE Extraction Buffer. Chromatin was extracted by 40 rounds of sonication 30s on, 30s off at 42% amplitude. Centrifugation was performed to separate soluble (supernatant containing chromatin) and insoluble (pellet of tissue debris) fractions.
After reverse cross-linking and proteinase K digestion, 25μl aliquots of each fraction were measured by a Qubit fluorometer (Invitrogen) and analyzed by agarose gel electrophoresis to check the quality and fragmentation of the extracted chromatin. Figure 2 shows a well-sheared chromatin with a good concentration (fragments are between 100-500 bp).-1.jpg)
Lane 1 – 100bp DNA ladder; Lane 2 – rat uterine FFPE chromatin, soluble fraction; Lane 3 – rat uterine FFPE chromatin, insoluble fraction; Lane 4 – human breast tumor FFPE chromatin, soluble fraction;
Lane 5 – human breast tumor FFPE chromatin, insoluble fraction.
|
FFPE Tissue
|
Soluble Fraction
|
Insoluble Fraction
|
|
Human breast tumor
|
6.68 ng/µL
|
3.44 ng/µL
|
|
Rat uterus
|
48 ng/µL
|
44 ng/µL
|
Figure 2. Agarose gel electrophoresis and Qubit measurements of chromatin extracted from rat and human FFPE tissue using the Chromatrap® FFPE ChIP kit.
Chromatin immunoprecipitation
Chromatrap® FFPE ChIP kit protocol was followed for the preparation of the immunoprecipitation slurries. In each 1ml slurry, 20 µL of chromatin stock was used with either 4 µg of anti-Histone H3 (Chromatrap® Product Code: 700000) for the positive immunoprecipitation or non-specific mouse IgG for the negative immunoprecipitation. Simultaneously, inputs were prepared containing 20 µL of the relevant chromatin stock; these were used for further analysis, and not subjected to ChIP enrichment.
The standard Chromatrap® FFPE ChIP protocol was adopted for carrying out immunoprecipitation. Slurries were incubated for one hour on an end to end rotator at 4 °C before loading onto the relevant Chromatrap® ProA ChIP column; this is followed by simple and quick centrifugation washes. After a 15 minute incubation of the FFPE Elution Buffer on the column, chromatin was eluted. Inputs and samples were reverse cross-linked for two hours, followed by Proteinase K digestion for one hour. Finally, the Chromatrap® FFPE purification columns and buffers were used for cleaning the samples, which were then eluted in 50 µL DNA Elution Buffer.
qPCR analysis
Primers were used to perform qPCR analysis of the rat or human GAPDH locus (Barber et al., 2005). Highly fragmented FFPE DNA can be efficiently analyzed using these primers as they generate amplicons <100bp. Analysis enabled precipitation efficiency dtermination and hence, specific enrichment at these gene loci in comparison with non-specific IgG. As a factor of the amount of input chromatin, the percentage of real signal was determined for the relative analysis between samples. The standard error of the mean of triplicate ChIPs was represented by error bars.
Results and discussion
The common epigenetic mark H3 was specifically enriched from chromatin extracted from FFPE human breast tumor tissue and rat uterine tissue to demonstrate the use of the Chromatrap® FFPE ChIP kit in the high yield extraction and excellent enrichment of chromatin from FFPE tissue.
The use of an antibody directed against H3 to enrich GAPDH locus in human chromatin (as shown in Figure 3) and animal chromatin (as shown in Figure 4) from FFPE tissue has resulted in outstanding signal to noise. The high positive antibody signal obtained from very low concentrations of chromatin illustrates the assay’s sensitivity, and the low non-specific binding shows its superior selectivity. Thanks to the versatility of the assay, better signal can be obtained from 100 ng of human breast tumor chromatin as well as 1µg of rat uterine chromatin.
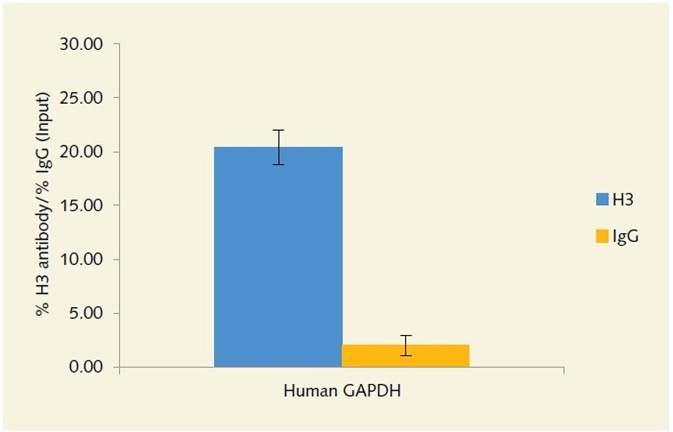
Figure 3. Enrichment of the GAPDH locus in human chromatin extracted from FFPE tissue,
using anti-histone H3 antibody.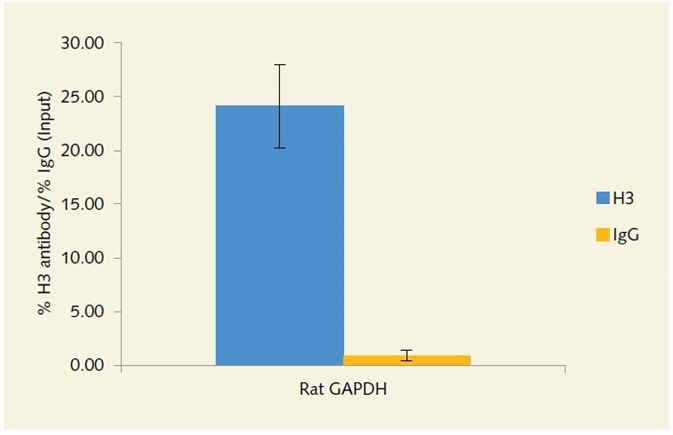
Figure 4. Enrichment of the GAPDH locus in rat chromatin extracted from FFPE tissue, using anti-histone H3 antibody.
Conclusion
Chromatin from complex FFPE tissue collected from animal and human sources can be efficiently extracted and immunoprecipitation performed using the Chromatrap® FFPE ChIP kit. A high yield of chromatin can be obtained from very complex sample sources using the extraction protocol and high real signal can be obtained from low concentration chromatin owing to the superior sensitivity of the unique solid state ChIP columns. These advantages together with high throughput capability and shorter protocols make the Chromatrap® FFPE ChIP kit a fast, versatile, reproducible and sensitive assay to analyze patient or research FFPE archives.
References
- Barber, R. D., Harmer, D. W., Coleman, R. A., Clark, B. J. (2005). GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiological genomics 21, 389-95.
- Beadling, C., Heinrich, M. C., Warrick, A., Forbes, E. M., Nelson, D., Justusson, E., Levine, J., Neff, T. L., Patterson, J., Presnell, A., McKinley, A., Winter, L. J., Dewey, C., Harlow, A., Barney, O., Druker, B. J., Schuff, K. G., Corless, C. L. (2011). J. Mol diagn 13:504-13.
- Bourgon, R., Lu, S., Yan, Y., Lackner, M. R., Wang, W., Weigman, V., Wang, D., Guan, Y., Ryner, L., Koeppen, H., Patel, R., Hampton, G. M., Amler, L. C., Wang, T. (2014). High-throughput detection of clinically relevant mutations in archived tumour samples by multiplexed PCR and next-generation sequencing. Clinical cancer research 20(8):2080-91.
- Fan, H., Gulley, M. L. (2001). DNA Extraction from Paraffin-Embedded Tissues. Molecular Pathology Protocols 49, 1-4.
- Fanelli, M., Amatori, S., Barozzi, I., Minucci, S. (2011). Chromatin immunoprecipitation and high-throughput sequencing from paraffin-embedded pathology tissue. Nat Protoc. 6(12):1905-19.
- Gilbert, M. T., Haselkorn, T., Bunce, M., Sanchez, J. J., Lucas, S. B., Jewell, L. D., Van Marck, E., Worobey, M. (2007). The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2(6):e537.
- Gillio-Tos, A., De Marco, L., Fiano, V., Garcia-Bragado, F., Dikshit, R., Boffetta, P., Merletti, F. (2007). Efficient DNA extraction from 25-year-old paraffin-embedded tissues: study of 365 samples. Pathology. 39(3):345-8.
- Lin, J., Kennedy, S. H., Svarovsky, T., Rogers, J., Kemnitz, J. W., Xu, A., Zondervan, K. T. (2009). High-quality genomic DNA extraction from formalin-fixed and paraffin-embedded samples deparaffinized using mineral oil. Anal Biochem. 395(2):265-7
- Pikor, L. A., Enfield, K. S., Cameron, H., Lam, W. L. (2011). DNA extraction from paraffin embedded material for genetic and epigenetic analyses. J Vis Exp. (49). pii: 2763.
- Rait, V. K., Zhang, Q., Fabris, D., Mason, J. T., O’Leary, T. J. (2006). Conversions of Formaldehyde-modified 2’-Deoxyadenosine 5’-monophosphate inc conditions modelling formalin-fixed tissue dehydration. J. Histochem Cytochem 54(3): 301-310.
- Su, Z., Dias-Santagata, D., Duke, M., Hutchinson, K., Lin, Y. L., Borger, D. R., Chung, C. H., Massion, P. P., Vnencak-Jones, C. L., Lafrate, A. J., Pao, W. (2011). A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small cell lung cancer. J. Mol diagn 13:74-84.
- Tang, W., David, F. B., Wilson, M. M., Barwick, B. G., Leyland-Jones, B. R., Bouzyk, M. M. (2009). DNA Extraction from Formalin-Fixed, Paraffin-Embedded Tissue. Cold Spring Harb Protoc, pdb prot5138 (2009).
About Chromatrap®
 Chromatrap® is a product of Porvair Sciences, a wholly owned subsidiary of Porvair plc.
Chromatrap® is a product of Porvair Sciences, a wholly owned subsidiary of Porvair plc.
They are one of the largest manufacturers of Ultra-Clean microplates, 96 well well filtration plates and Microplate handling equipment for life science and synthetic chemistry. With offices and Class VIII clean room manufacturing located in the UK, combined with a world-wide network of distributors and dedicated distribution hub in the USA, we pride ourselves on their continuous innovation, research and flexibility to meet customer demands. We offer OEM production and contract manufacturing through their North Wales facility.
Their porous polymeric material, BioVyon™, whose chemical functionalisation can endow it with internal surface properties individually configured to capture and separate target species out of difficult mixtures, has opened up many possibilities in the field of BioSciences where molecules of interest such as DNA, RNA, proteins etc can be selectively pulled out of complex mixtures of biological origin. The materials have proven to be a remarkably good substrate for accepting novel chemistries such as the organically bound Protein A and Protein G in Chromatrap®.
Using their 25 years experience of microplate manufacturing, Porvair Sciences has now developed a high-throughput bead-free ChIP assay based on their filtration plates containing their Chromatrap chemistry. Chromatrap-96 enables large scale epigenetic screening to become a reality in many laboratories and eliminates many of the long and laborious steps previously undertaken in such work.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.