This article describes how the Dental Forensic Extraction Kit (DFKMR), InnoQuant® HY DNA Quantification Kit, and the MiSeq FGx™ Forensic Genomics System can be used to quickly gather conclusive results from complex samples.
Introduction and Methods
Degraded DNA presents a serious limitation for forensic DNA laboratories and is frequently found in cold case evidence, crime scene samples, and unidentified remains related to tragedies from mass disasters or missing persons cases.
Highly degraded samples, for example, bones and teeth, are conventionally assigned to mitochondrial DNA processing because of the insufficient results attained with regular nuclear DNA testing standards.
The latest advancements in extraction techniques performed upstream and quantification chemistries, along with massively parallel sequencing (MPS) technology, all enhance the chances of gathering data-intensive nuclear DNA profiles from complex samples.
The data outlined in this article shows the extraordinary performance of the thorough workflow used on the degraded DNA samples. This workflow comprises the following:
- Dental Forensic Kit (DFK) – An original technique for the recovery of dental tissue produced at Universidad de los Andes, Santiago, Chile (Figure 1) [1,2,3,4,5]
- InnoQuant HY DNA Quantification Kit (InnoGenomics) – A highly sensitive real-time qPCR assay for establishing the integrity, quality, and quantity of human and male DNA at the same time (Table 1) [6,7]
- MiSeq FGx Forensic Genomics System (Verogen) – A DNA-to-data MPS solution created exclusively for applications within forensic DNA (Figure 2) [8,9,10,11,12]
The main benefits of each technique include:
- Dental Forensic Kit (DFK)
- A more efficient extraction with greater yield
- Excluding the conventional grinding step mitigates the risk of contamination
- The tooth is preserved to enable future dental examinations or re-extraction
- InnoQuant HY DNA Quantification Kit
- Highly reproducible and sensitive DNA quantitation down to less than 0.001 ng/µL utilizing high copy number retrotransposable element (RE) targets.
- Optimized genotyping success rate through precise degradation index, true negative screening, and PCR inhibitor detection.
- MiSeq FGx Forensic Genomics System
- Amplification of up to 230 STR and SNP markers at the same time, along with Amelogenin and 200 amplicons with a range of 63 to 180 bp in size. [10]
- Profiles that are highly discriminating can be repeatedly acquired with ≤ 16 pg total DNA input. [9]
- Able to produce biogeographical ancestry and phenotype estimation all at once. [9,10,11]
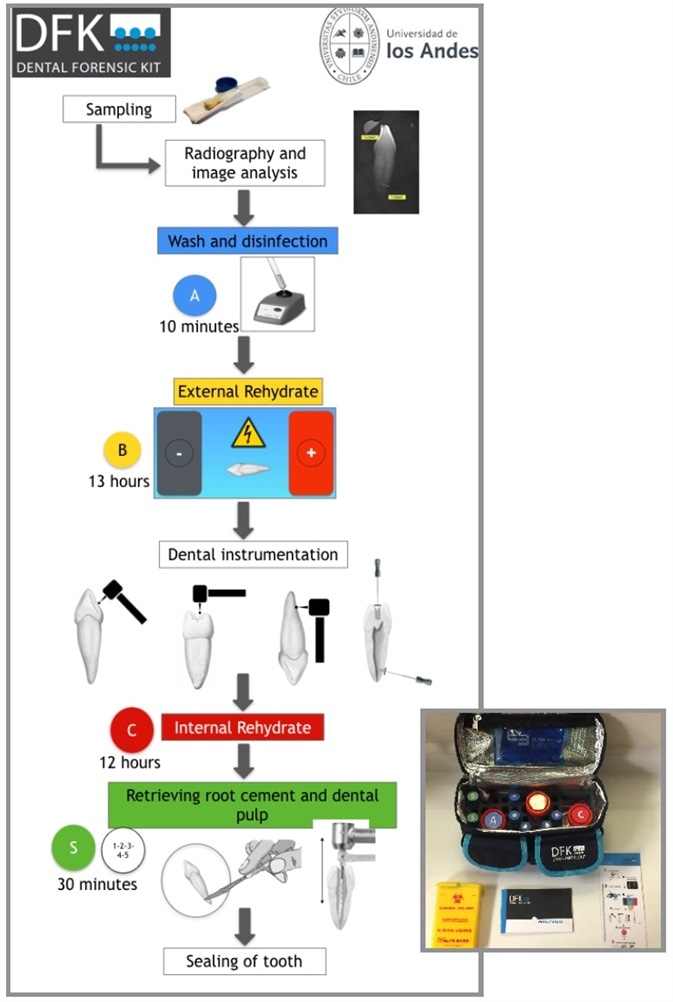
Figure 1. DFK method overview (Universidad de los Andes, Santiago, Chile). Image Credit: Verogen, Inc.
The DFK method enhances DNA recovery from dental remains with an original and fast dental tissue extraction technique.
This dynamic technique can additionally be applied to bone samples. The system starts with radiographic imaging of the tooth, with washing and external rehydration being performed after this. A specific dental instrument then perforates the bone or tooth to enable internal rehydration while reducing damage to the sample.
Both dental pulp and root cement can be acquired for resulting DNA extraction employing the Quick Extract™ FFPE DNA Extraction Kit (Lucigen®). The tooth is then sealed and retained, enabling future testing if required. [1,2,3,4,5]
Degradation Study Experimental Design
To assess the performance of InnoQuant HY quantification and MPS analysis in combination with the MiSeq FGx System, a degradation investigation was first carried out using DNA sourced from an anonymous male blood bank donor.
Levels of degradation from moderate to severe were acquired using sonication at 50 °C varying from zero to 16 hours. Utilizing InnoQuant HY, DNA extracts were quantified to gather long and short target concentrations (ng/µL) and the related degradation index (DI = [Short/Long]) for every sample.
From the quantified DNA, sequencing libraries were set up employing the ForenSeq DNA Signature Prep Kit, using a maximum DNA input volume of 5 µL. Sequencing on the MiSeq FGx Instrument and data analysis utilizing the ForenSeq Universal Analysis Software was then performed.
After the primary degradation investigation with artificially degraded DNA, a jaw bone specimen, and several teeth with postmortem intervals (PMI) varying from seven days to 45 years were analyzed.
The Universidad de los Andes in Santiago, Chile performed a collaborative experiment using the DFK extraction technique to extract the dental pulp, root cement, and jaw bone DNA. The InnoQuant HY was then used to quantify the samples before processing with the MiSeq FGx Forensic Genomics System.
Table 1. InnoQuant HY Kit Configuration. Source: Verogen, Inc.
| Target |
Genomic Location |
Amplicon Length |
Reporter Dye |
| Short |
Yb8 autosomal RE |
80 |
HEX |
| Long |
SVA autosomal RE |
207 |
Cy5 |
| Male |
Y chromosome |
79 |
FAM |
| IPC |
Synthetic sequence |
172 |
TAMRA |
Table 1: InnoQuant HY Kit Configuration - InnoQuant HY is a live qPCR system consisting of four targets of DNA – two RE autosomal targets of different sizes, an internal positive control (IPC) target, and male-specific targets.
The use of two autosomal targets strongly enhances the reproducibility and sensitivity of the quantitation of DNA and the identification of degradation in forensic samples, these are high copy number Alu and SVA Res (greater than 1,000 copies per genome). [6,7]

Figure 2. The MiSeq FGx Forensic Genomics System. Image Credit: Verogen, Inc.
The MiSeq FGx System includes the MiSeq FGx Instrument, the ForenSeq Universal Analysis Software, and the ForenSeq DNA Signature Prep Kit.
Magnetic bead-based chemistries and PCR amplification are used in the ForenSeq DNA Signature Prep Kit to construct targeted DNA sequencing libraries for the MiSeq FGx instrument.
MPS data are produced for up to 230 genetic markers along with Amelogenin per sample. Once sequencing has been completed, the ForenSeq Universal Analysis Software creates semi-automated allele calls and presents genotype data for the analyst to observe.
Along with short tandem repeats (X chromosome, Y chromosome, and autosomal STRs), single nucleotide polymorphism (SNP) markers are targeted, such as phenotypic-informative SNPs (piSNPs), biogeographical ancestry-informative SNPs (aiSNPs), and identity-informative SNPs (iSNPs). [8,9,10,11,12]
Results
The blood samples that had been degraded progressively produced long target concentrations ranging from 0.87 to 0.0004 ng/µL and short target concentrations from 0.87 to 0.17 ng/µL.
The degradation index (DI) varied from 1 (no identified degradation) to 460 (strongly degraded). The samples were amplified with ForenSeq in line with the InnoQuant HY long target quantities, producing total DNA inputs varying from 1.7 to 0.002 ng.
Figure 3 presents the total number of STR alleles called and iSNP loci typed for samples with progressing degrees of degradation.
The sum of autosomal, Y- and X- STR alleles generated for every sample varied from 85 (100%) with a DI of 1 at 1 ng total DNA input to 23 alleles called (27%) with a 460 DI and 2 pg of total DNA input. The sum of all iSNP loci typed varied from 94 to 70 (100% to 74%).
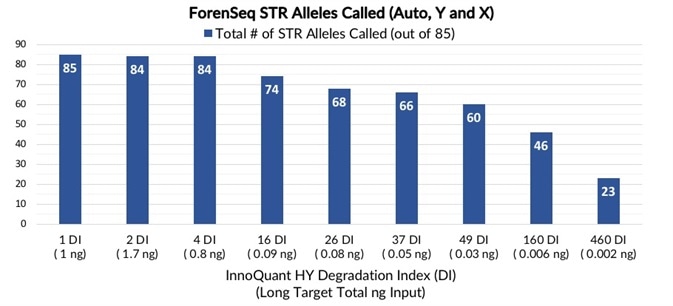
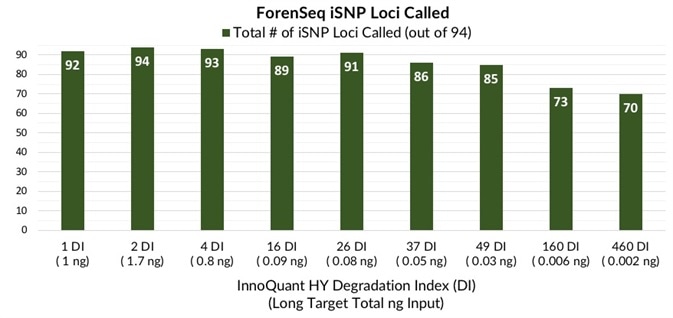
Figure 3. Total number of STR alleles and iSNP loci - Total number of STR alleles detected (blue) and total number of iSNP loci called (green) for artificially degraded blood samples with DI ranging from 1 (not degraded) to 460 (severely degraded). Image Credit: Verogen, Inc.
Along with iSNP and STR data, piSNP and aiSNP data offers the biogeographical ancestry estimation and the phenotype in the ForenSeq Universal Analysis Software.
Estimations of the hair and eye color phenotype, which demand 100% piSNP locus call rates, were effectively produced for the initial four samples down to 90 pg total input and a DI of 16 [11].
Estimations of hair color for the four samples sourced from the same anonymous blood donor gave the following suggestions: 74% red, 22% blond, 4% brown and 0% black.
Estimations of eye color were 96 to 97% blue, 2 to 3% intermediate, and 1 to 2% brown. The prediction of biogeographical ancestry was repeatedly shown as the European population group for each of the nine samples (shown in Figure 4).
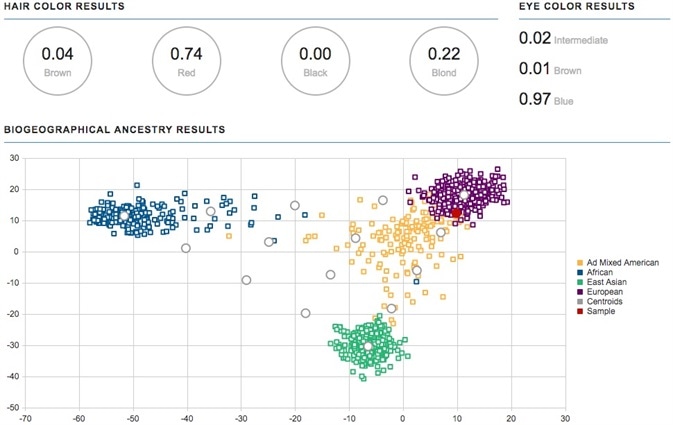
Figure 4. Phenotype and biogeographical ancestry estimation - Hair color, eye color and biogeographical ancestry estimation in the ForenSeq Universal Analysis software for degraded blood with 90 pg input and DI of 16 (sample indicated with red dot). Image Credit: Verogen, Inc.
In the investigation of the dental remains, the data suggested that the dental pulp or root cement extracted utilizing the DFK technique generated DNA of adequate quantity and quality for most teeth samples.
Multiple extracts with PMIs varying from seven days to around six months were not degraded to a significant extent.
Figures 5 and 6 show that dental extracts with DI varying from 0.7 to 7.8 and long target total DNA inputs from 0.05 to 5.7 ng caused 58 to 94 iSNP loci typed and 46 to 49 ForenSeq autosomal STR allele calls.
In comparison, the equivalent dental extracts previously produced 25 to 31 autosomal STR allele calls with Identifiler® Plus amplification utilizing the maximum 10 µL DNA input run on the Applied Biosystems® 3100 CE instrument.
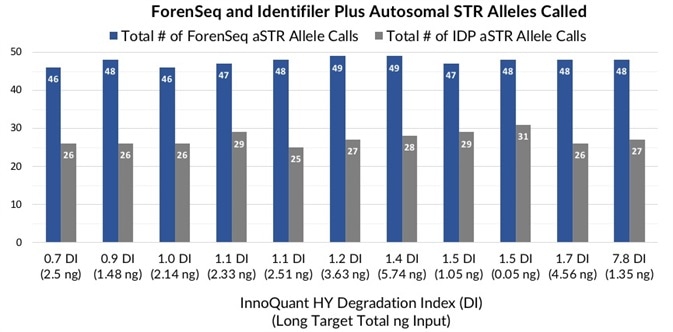
Figure 5. Total number of ForenSeq (blue) and Identifiler Plus (grey) autosomal STR alleles detected in teeth extracts with DI ranging from 0.7 to 7.8. Image Credit: Verogen, Inc.
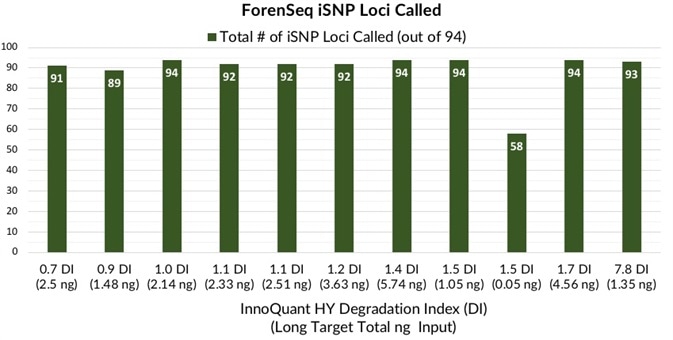
Figure 6. Total number of ForenSeq iSNP loci detected in teeth extracts with DI ranging from 0.7 to 7.8. Image Credit: Verogen, Inc.
Highly discriminating SNP and STR results, very low levels of DNA input, and an indication of inhibition were identified by the InnoQuant HY IPC in the more challenging samples, even though a higher degree of degradation was observed.
One of the more difficult teeth samples with a total long target input of 0.001 ng and a DI of 76 generated 78 STR alleles in total and 85 typed iSNP loci (Figure 7).
A 45-year PMI jaw bone extract was also effectively typed with 59 iSNP loci and 48 STR alleles called at a DI of 4.7 and a long target input of 0.01 ng (Figure 7). This 45-year PMI bone sample had generated no STR alleles before with the Identifiler Plus kit run on the 3100 instrument.
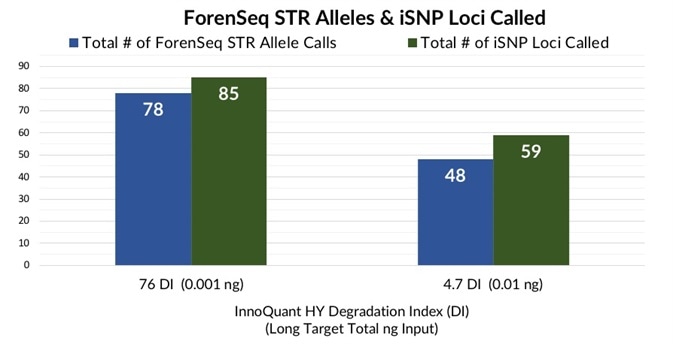
Figure 7. Total number of ForenSeq STR alleles and iSNP loci detected in extremely low quantity/quality teeth and bone extracts. Image Credit: Verogen, Inc.
Conclusions
The combined DFK extraction technique, InnoQuant HY quantification, and MiSeq FGx Forensic Genomics System offers a powerful workflow enabling the user to acquire effective nuclear DNA results from complex, degraded, and low-level samples.
DFK extraction allowed an adequate amount of DNA to be recovered, and InnoQuant HY offered precise quantification data to allow for optimal amplification with the ForenSeq DNA Signature Prep Kit. Applying this system to real-life, complex dental remains produced a high amount of viable data for identification.
References and Further Reading
- Universidad de los Andes, Santiago, Chile DFK Patent: https://patentscope.wipo.int/search/en
- http://www.lucigen.com/home.php?cat=282
- P. Carrasco, C. Brizuela, I. Rodriguez, S. Muñoz, M. Godoy, C. Inostroza, Histological transformations of the dental pulp as possible indicator of post mortem interval: a pilot study. https://doi.org/10.1016/j.forsciint.2017.09.001
- P. Carrasco, C. Inostroza, M. Godoy M, G. Gak, Innovative method and kit DFK(MR) of sample preparation from dental human remains for human genetic identity, Forensic Science International Genetics (fsigen 2018-7), Submitted January 12, 2018.
- Application of NGS in the analysis of human DNA from dental remains with new Forensic Kit DFK(MR) Unpublished results (manuscript in preparation)
- http://innogenomics.com/
- A. Loftus, G. Murphy, H. Brown, A. Montgomery, J. Tabak, J., Baus, et al., Development and validation of InnoQuant® HY, a system for quantitation and quality assessment of total human and male DNA using high copy targets, https://doi.org/10.1016/j.fsigen.2017.04.009
- http://www.verogen.com/
- A. Jager, M. Alvarez, C. Davis, E. Guzman, Y. Han, L. Way, et al., Developmental validation of the MiSeq FGx Forensic Genomics System for targeted next generation sequencing in forensic DNA casework and database laboratories Forensic Sci. Int. Genet. 28 (2017) 52-70. https://doi.org/10.1016/j.fsigen.2017.01.011
- ForenSeq DNA Signature Prep Reference Guide http://www.verogen.com/
- ForenSeq Universal Analysis Software Guide v1.2 http://www.verogen.com/
- MiSeq FGx Instrument Reference Guide http://www.verogen.com/
About Verogen, Inc.
Introducing the world’s first sequencing company solely dedicated to forensic science. Forensic laboratories are unique, and require unique scientific solutions.
We get this. Because we live it. We create thoughtfully tailored genomic solutions for forensic DNA labs. The kinds of tools we longed for when we worked in those labs ourselves. We formed an independent company with dedicated resources because we believe it’s the best way to fully address those unique needs.
The last few decades have seen tremendous advancements in molecular biology techniques, such as massively parallel sequencing. While our colleagues in other scientific fields have embraced and benefited from these improvements, forensic genomics has yet to realize the potential of the evolved technology. It’s time we had modern tools tailored to our needs.
Verogen understands this. Our sole focus is to advance science to help unlock the true potential of forensic genomics. Powered by Illumina technology and free of legacy method allegiance, we are uniquely positioned to support forensic labs with innovative solutions purpose-built for the challenges of DNA identification. Working in partnership with the community, we can elevate the forensic genomics lab’s role in preserving public safety—and improve global justice for all.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.