One of the most common themes associated with all omics applications is the ever-increasing size of patient cohorts. This increase is prompted by the need to identify novel disease biomarkers and an increase in study power, but throughput becomes the limiting factor as studies scale to incorporate thousands of patient samples.
Methods such as Rapid Microbore Metabolic Profiling (RAMMP) can decrease separation time by scaling down column dimensions and chromatographic separation time to improve throughput.1 These improvements can, however, come at the cost of feature detection and peak capacity.
It is possible to maintain narrow peak widths by reducing post-column volume and moving the column closer to the source. This approach can be employed in conjunction with a vacuum jacket to mitigate frictional heating and temperature differentials. These vacuum jacketed columns (VJC) can increase peak capacity considerably while narrowing peak widths.2
High-resolution mass spectrometers capable of rapid scanning with no loss of resolution are required to reliably identify lipids in short analysis times. Multi-reflecting time of flight leverages gridless mirrors to prevent ion loss while increasing flight length, and therefore, mass resolution.
For example, the benchtop Xevo™ MRT Mass Spectrometer can scan up to 100 Hz while maintaining a mass resolution of up to 100,000 full width half maximal (FWHM). These capabilities are essential for sufficiently profiling ever-decreasing peak widths (Figure 1).
Therefore, vacuum jacketed columns and the Xevo™ MRT MS were used to analyze the lipidome of healthy controls and patients with various cancers. These results were then compared with traditional chromatography.
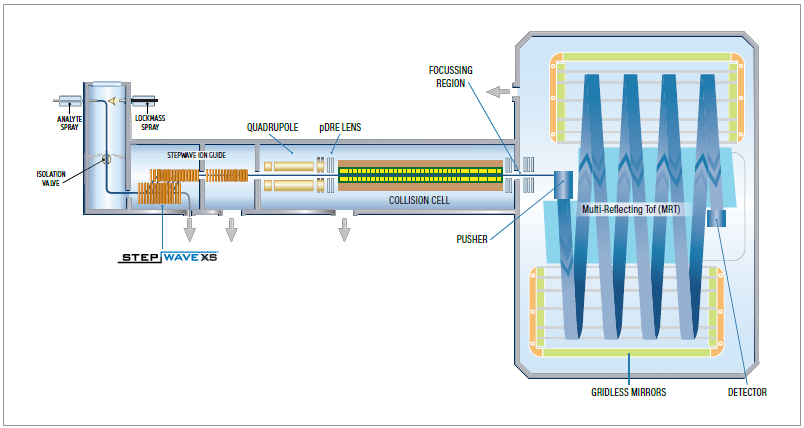
Figure 1. Schematic of the Xevo MRT MS. Image Credit: Waters Corporation
Methods
Lipids from serum samples of patients with colon or rectum cancer and healthy controls were extracted via IPA before being spiked with EquiSPLASH™ Standard at 100 ng/ml to act as an internal control.
Extracted lipids were initially separated on a 2.1 mm x 50 mm diameter CSH™ UPLC™ column over a 4-minute gradient at 0.8 ml per minute.
Solvent A was ACN:H2O:1M aqueous ammonium formate (600:390:10 v/v), and Solvent B was IPA:ACN:1M aqueous ammonium formate (900:90:10 v/v).
The gradient started at 1 % B before a linear increase to 30 % B was completed at 0.1 minutes before 90% B at three minutes. Column wash at 99.9 % B was done at 3.2 minutes for a total of 0.5 minutes prior to column re-equilibration at 1 % B until
4 minutes. An additional, corresponding column in VJC format (2.1 mm x 50 mm CSH) was applied for the separation.
The flow rate for VJC configurations was scaled down to 0.5 ml per minute while maintaining a chromatographic gradient over 4 minutes. A shortened equivalent gradient of 2 minutes and 1 minute, respectively, was then applied.
The eluate was directed towards the Xevo™ MRT MS, which was operating at a scan speed of 20 hz (4-minute and 2-minute gradient) or 30 hz (1-minute gradient).
Data was collected using a data-independent mode of acquisition (MSE) with alternate scans of high collision energy (20-40 eV ramp) and low collision energy (6 eV).
Generated data was converted to the mzML format via the DATA Convert application. This was done as the injection finished acquiring, prior to import into the Lipostar2 software from Mass Analytica. This enabled peak picking, identification of features, and identification of lipids using the LIPID MAPS® structure database.3
Additional analysis of exported compound measurements from Lipostar2 was done using MetaboAnalyst (Version 6). This enabled the generation of PLS-DA plots.
The LC-MS Toolkit application analyzed retention time and chromatographic peak width. This application is available via the waters_connect™ software platform.
Results
Initial experiments using a conventional UPLC™ column highlighted a complex chromatogram as anticipated, with clear and observable differences between the cancer samples and healthy controls when PLS-DA was applied to the data.
Switching the configuration to a VJC format while maintaining gradient time also revealed differences between the groups. However, this approach displayed a much greater number of features and subsequently identified lipids.
There was a notable reduction in base peak width of chromatographic peaks when using a conventional column versus VJC (Figure 2) and an increase in peak height resulting from increased ionization efficiency from a lower flow rate.
This trend was observed irrespective of the compound’s retention time. Figure 2 shows the elution of phosphatidylserine, one of the earliest eluting compounds during lipid analysis.
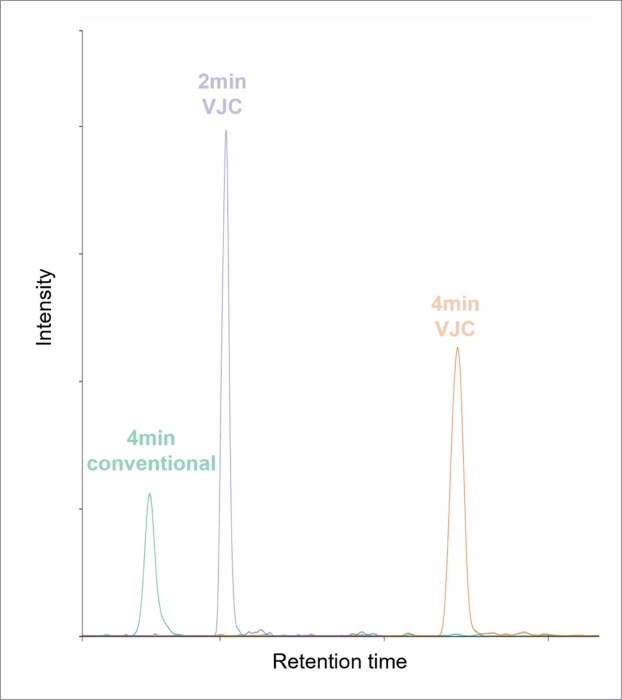
Figure 2. Extracted ion chromatogram of phosphatidylserine with m/z 755.5562 using a 2.1 mm conventional column (green), 2.1 mm VJC (4 min method; orange) or 2.1 mm VJC (2 min method; blue). Image Credit: Waters Corporation
Scaling down to 2 minutes of gradient time with VJC was found to further decrease base peak width, leading to a corresponding increase in peak height. Increased peak height also resulted in improved signal-to-noise using VJC, therefore improving the sensitivity of this method.
This increase in sensitivity allowed 3.5 times the number of features to be identified using VJC when analyzing cancer patient samples compared to a traditional column (Figure 3).
The applicability of a shorter gradient was investigated to continue reducing analysis times. Shortening the gradient by 50% using a VJC was also found to yield more features versus a 4-minute method with a traditional column. This led to a 100% increase in throughput and an approximately 70% reduction in solvent consumption.
The decrease in gradient time had no effect on the method’s ability to sufficiently profile the chromatographic peak. This was due to the Xevo™ MRT MS’s rapid scanning capabilities. There was also no impact on mass resolution, and no loss of diagnostic power was noted.
The analysis of a full study cohort of patients with colorectal cancer versus healthy controls showed clear separation using PLS-DA. Study QCs were accurately located on the plot in this study.
PLS-DA again showed clear differences between the control versus disease group when utilizing a 1-minute VJC method. This approach afforded the study a 300% increase in throughput and an approximately 85% decrease in solvent consumption versus the 4-minute method with a traditional column.
Discussion
There is increasing pressure on analysts to improve throughput, posing ongoing challenges to data quality maintenance. There is also a drive towards analysis methods that are more environmentally friendly by lowering their consumption of organic solvents.
Using vacuum-jacketed columns in conjunction with the high resolution and rapid scanning capabilities of the Xevo™ MRT MS offers unrivaled throughput with no compromise in data quality.
Transferring a rapid 2.1 mm conventional column method to a VJC format offered 3.5 times improved feature detection. An increase in peak capacity via VJC also reduced flow rate and, therefore, solvent consumption. This provided better results than conventional chromatography and offered a more environmentally friendly solution.
Depending on the final purpose of the analysis, methods can be scaled down for increased throughput or kept the same should a deeper investigation into the lipidome be required. Plumb et al. (2022) showed that VJC performed better in every metric measured versus a conventional column.
As pressure to deliver increased throughput continues, it is vitally important to use a mass spectrometer with a scan speed able to sufficiently profile ever-decreasing chromatographic peaks.
The Xevo™ MRT MS has successfully demonstrated that capability without sacrificing resolution, making it ideally suited for use in high-throughput studies.
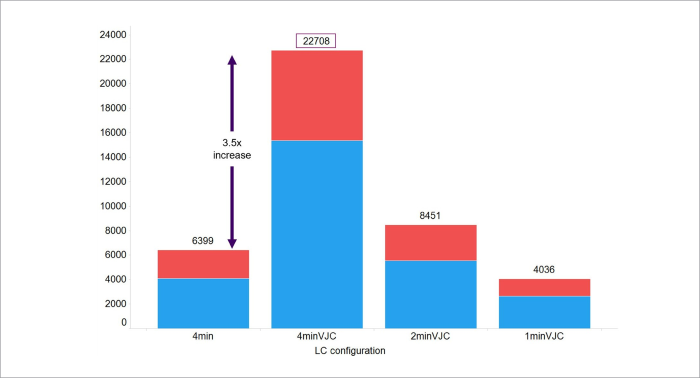
Figure 3. Stacked bar chart representing identified lipids (red) and unidentified features (blue) found using four different column and gradient combinations. Image Credit: Waters Corporation
Conclusion
The usefulness and suitability of VJC for the high-throughput analysis of lipids have been shown using cancer patient samples. Transferring a standard conventional column method to VJC resulted in 3.5 times more features being identified and an approximately 37% reduction in solvent consumption.
It was also noted that halving the gradient time while maintaining the flow rate led to identifying more features than a longer conventional method. This approach also resulted in the use of around 70% less solvent.
It was observed that the use of an ultra-fast 1-minute VJC method demonstrated sufficient profiling of the study cohort and clear separation between the healthy and disease states via PLS-DA.
The Xevo™ MRT MS was shown to offer excellent and uncompromising mass resolution. When combined with rapid scanning, these capabilities facilitated the identification of many features from profiling narrow chromatographic peaks. This feat has not been historically possible using trapping mass spectrometers.
References and further reading
- Gray, N., et al. (2016). Development of a Rapid Microbore Metabolic Profiling Ultraperformance Liquid Chromatography–Mass Spectrometry Approach for High-Throughput Phenotyping Studies. Analytical Chemistry, 88(11), pp.5742–5751. https://doi.org/10.1021/acs.analchem.6b00038.
- Plumb, R.S., et al. (2021). High Throughput UHPLC-MS-Based Lipidomics Using Vacuum Jacketed Columns. Journal of Proteome Research, 21(3), pp.691–701. https://doi.org/10.1021/acs.jproteome.1c00836.
- Conroy, M.J., et al. (2023). LIPID MAPS: update to databases and tools for the lipidomics community. Nucleic Acids Research. https://doi.org/10.1093/nar/gkad896.
- Pang, Z., et al. (2024). MetaboAnalystR 4.0: a unified LC-MS workflow for global metabolomics. Nature Communications, (online) 15(1), p.3675. https://doi.org/10.1038/s41467-024-48009-6.
Acknowledgments
Produced from materials originally authored by Matthew E. Daly, Nyasha Munjoma, Rob S. Plumb, Jason Hill, Nick Tomczyk, Lee A. Gethings, and Richard Lock from Waters Corporation.
About Waters Corporation
At Waters, we unlock the potential of science by solving problems that matter. Our software and instruments ensure the safety of the medicines we take, the purity of the food we eat and the water we drink, and the quality and durability of products we use every day. Together with our customers, in labs around the world, we deliver scientific insights to improve human health and well-being, helping to leave the world better than we found it.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.