The Xevo™ MRT Mass Spectrometer boasts unparalleled performance and speed, offering users high-resolution time-of-flight data that is wholly independent of spectral acquisition rate.
The instrument’s impressive MS specification includes 100,000 (FWHM) resolution at up to 100 Hz scan rates, in conjunction with <500 ppb mass accuracy. Analytes can be identified confidently at throughput levels suitable for large cohort experiments.
The pioneering multi-reflecting ToF technology from Waters™ Corporation’s SELECT SERIES™ MRT has now been scaled into a compact benchtop platform designed to help address the most challenging epidemiological studies and biomedical research issues.
Performance is critical in large-scale metabolic phenotyping experiments, from measurement precision to instrument stability over large studies. These factors are key to generating high-quality experimental outcomes.
The Xevo™ MRT’s novel acquisition system features a powerful dual-gain analog-to-digital converter, ensuring long-term system stability over a high dynamic range and high-quality data with consistent reproducibility and robustness.
This article outlines the use of an exemplar sample set to highlight the Xevo™ MRT Mass Spectrometer’s suitability for analyzing colorectal cancer human serum samples.1
These capabilities are combined with market-leading chemistry and separations, premier technology, and advanced informatics to create a complete lipidomic workflow, featuring the LipoStar2 from Mass Analytica.
Experimental conditions
Samples
Source: Waters Corporation
| Samples |
Colorectal cancer (CRC)
human serum samples |
6 Healthy control plasma |
| 4 Colon cancer plasma |
| 2 Rectum cancer plasma |
| NIST SRM 1950 plasma |
| Study Reference/pooled (QC) |
| A 100x dilution of EquiSPLASH |
Source: Waters Corporation - Forensics
| LC conditions |
| LC System |
Waters ACQUITY™ Premier |
| Column |
ACQUITY Premier UPLC™ CSH™
C18 2.1 x 50 mm, 1.7 µm |
| Column Temperature |
55 °C |
| Sample temperature |
8 °C |
| Injection volume |
0.5 µL |
| Mobile phase A |
50:25:25 H20:IPA:MeCN
w/5 mM ammonium acetate and 0.05% acetic acid |
| Mobile phase B |
50:50 IPA:MeCN
w/5 mM ammonium acetate and 0.05% acetic acid |
| Flow rate |
0.4 mL/min |
Source: Waters Corporation
| Gradient table |
| Time (min) |
Flow (mL/min) |
%A |
%B |
Curve |
| Initial |
0.8 |
99.0 |
1.0 |
Initial |
| 0.05 |
0.8 |
70.0 |
30.0 |
6 |
| 3.00 |
0.8 |
10.0 |
90.0 |
6 |
| 3.20 |
0.8 |
0.1 |
99.9 |
6 |
| 3.70 |
0.8 |
0.1 |
99.9 |
1 |
| 4.00 |
0.8 |
99.0 |
1.0 |
1 |
Source: Waters Corporation - Forensics
| MS conditions |
| Instrument |
Xevo MRT Mass Spectrometer |
| Acquisition |
Electrospray positive and negative ion mode |
| Capillary voltage |
2.8 V/2.0 V |
| Desolvation temperature |
500 °C |
| Source temperature |
120 °C |
| Cone |
40 V |
| Mass range |
m/z 50–1200 |
| Acquisition rate |
10 Hz (for 200 injections) and 1, 5, 10 and 20 Hz |
| Acquisition mode |
MSE |
| Collision ramp in HE |
20–45 eV |
Acquisition and
processing software |
waters_connect™ platform with Sample Sub, AME and DATA Convert applications. LipidStar2, Mass Analytica |
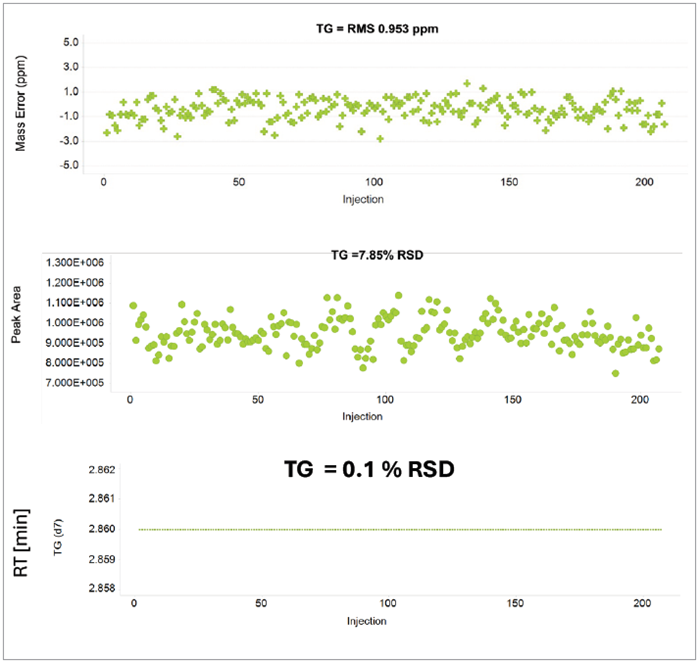
Figure 1. Mass measurement accuracy and peak area over 200 injections of a triglyceride lipid spiked into Plasma. Image Credit: Waters Corporation
Results
In the example presented here, the Xevo™ MRT Mass Spectrometer was used to analyze a sample set of more than 200 injections based on the Rapid Microbore Metabolic Profiling (RAMMP) methodology.2
This approach offered excellent analytical reproducibility in terms of mass accuracy, retention time, and peak area, showcasing the system’s suitability for complex epidemiological studies or biomedical research.
Colorectal cancer human serum samples were prepared in duplicate before being injected in triplicate in a randomized order. EquiSPLASH was spiked into the NIST plasma, and the study QC samples were injected every six sample injections.
It was noted that throughout 200 injections, deuterated triglyceride d7 was observed with a peak area measurement of 7.8% RSD and a mass accuracy of 0.95 ppm RSD (Figure 1).
The excellent mass accuracy demonstrated by these results allows analysts to use tight tolerances when performing compound identification, effectively lowering turnaround time from analysis to result.
High-quality statistical analysis was made possible thanks to the exceptional tolerances on retention time and peak area measurement of 0.1% and 7.8% RSD, respectively. These tolerances were successfully demonstrated within Mass Analytica’s LipoStar2 software.
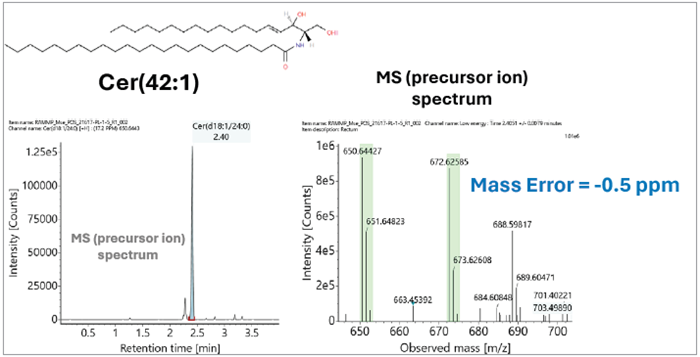
Figure 2. Identification of the ceramide lipid in the CRC study. Image Credit: Waters Corporation
LipoStar2 has been designed to perform the statistical analysis required to establish sample groupings and identify lipids, facilitating high-quality scientific interpretation via database searching and pathway profiling.3
In the example presented here, LipoStar2’s pathway profiling functionality enabled identification of the endogenous ceramide lipid Cer 18:1/24:0 (Figure 2) within the colon versus rectum cancer sample sets. This is an upregulated lipid in CRC dysregulation.4
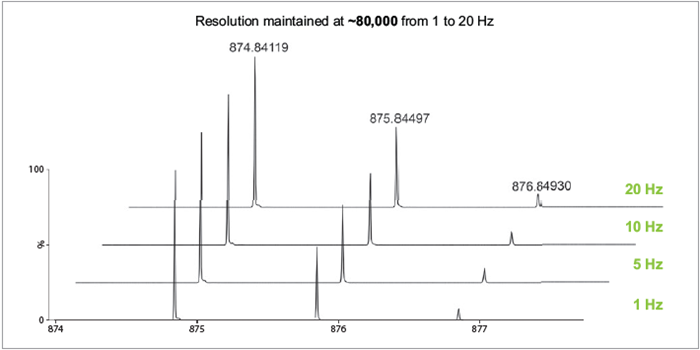
Figure 3. Mass resolution of 80,000 FWHM maintained at scan rates of 1, 5, 10, and 20 Hz. Image Credit: Waters Corporation
High-quality data generated from multi-reflecting time-of-flight technology offers consistent mass resolution over a broad range and is independent of acquisition rate. The capabilities were demonstrated on the endogenous lipid TG(52:3), which was successfully measured at 1, 5, 10, and 20 Hz at a resolution of 80,000 FWHM (Figure 3).
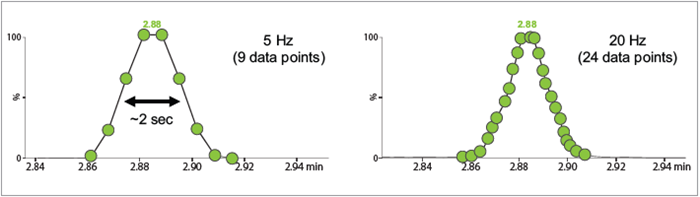
Figure 4. Extracted ion chromatogram of a triglyceride lipid from a 4 min LC/MS acquisition measured at 5 and 20 Hz. Image Credit: Waters Corporation
The system is capable of performing both qualitative and quantitative analysis. Figure 4 displays the effect of acquisition rate on the number of points across at the modest scan speed of 20 Hz and the lower rate of 5 Hz.
The Xevo™ MRT Mass Spectrometer has the potential to acquire at more rapid scan speeds, enabling increased throughput and lab productivity. It can be used at up to 50 Hz in MS mode and 100 Hz in MS/MS mode.
Summary
The Xevo™ MRT Mass Spectrometer’s exceptional system performance and reproducibility enabled the performance of an extensive lipidomic study, highlighting the importance of system stability when evaluating a range of samples’ statistically significant biological differences.
Using the waters_connect™ platform applications, it was possible to acquire and export samples via .mzML. This enabled their data processing, interpretation, and library searching via LipoStar2.
The study presented here showcases a high-throughput lipidomic workflow that combines column chemistries, separations, and informatics with the extraordinary data quality of the Xevo™ MRT Mass Spectrometer, ultimately empowering scientists to make meaningful scientific discoveries.
References and further reading
- Morgan, E., et al. (2022). Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut, (online) 72(2), p.gutjnl-2022-327736. https://doi.org/10.1136/gutjnl-2022-327736.
- Waters (2018). Rapid Microbore Metabolic Profiling (RAMMP) with Ion Mobility for the Lipidomic Investigation of Plasma from Breast Cancer Patients. (online) Available at: https://www.waters.com/nextgen/in/en/library/application-notes/2018/rapid-microbore-metabolic-profiling-ion-mobility-lipidomic-investigation-plasma-breast-cancer-patients.html?srsltid=AfmBOopeMSO2QwOG-bQeRpqDaWTNrqjNjepbizjniwCtXy-RjFblqoVc (Accessed 23 Apr. 2025).
- Goracci, L., et al. (2017). Lipostar, a Comprehensive Platform-Neutral Cheminformatics Tool for Lipidomics. Analytical Chemistry, 89(11), pp.6257–6264. https://doi.org/10.1021/acs.analchem.7b01259.
- Gomez-Larrauri, A., et al. (2021). Ceramide Metabolism Enzymes-Therapeutic Targets against Cancer. Medicina (Kaunas, Lithuania), (online) 57(7), p.729. https://doi.org/10.3390/medicina57070729.
Acknowledgements
Produced from materials originally authored by Waters Corporation.
About Waters Corporation
At Waters, we unlock the potential of science by solving problems that matter. Our software and instruments ensure the safety of the medicines we take, the purity of the food we eat and the water we drink, and the quality and durability of products we use every day. Together with our customers, in labs around the world, we deliver scientific insights to improve human health and well-being, helping to leave the world better than we found it.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.