Concept Life Sciences recently developed BioPALS, an innovative, lean, and versatile hit identification platform that leverages a combination of AI-powered virtual screening and pioneering biophysical technologies to facilitate the rapid identification and comprehensive characterization of hits.
Image Credit: Concept Life Sciences
This article looks at the implementation of BioPALS technology with the BRPF1b bromodomain, using BioPALS to identify a range of selective, potent, and lead-like hits that present new opportunities for the treatment of cancers such as AML.
BRPF1b – An actionable target
Bromodomains are epigenetic reader proteins that recognize acetylated histone tails, playing a key role in chromatin remodeling and gene regulation. Through these mechanisms, they influence the expression of genes involved in cancer, inflammation, and neurological disorders.1
One such protein, Bromodomain and PHD finger-containing protein 1b (BRPF1b), regulates transcription through multiple chromatin reader domains—including a double PHD and zinc finger assembly, a bromodomain, and a C-terminal PWWP domain.
This multi-domain structure enables BRPF1b to coordinate complex epigenetic signaling events, making it an attractive therapeutic target.
BRPF1b has been specifically linked to diseases such as **hepatocellular carcinoma (HCC)**2 and **acute myeloid leukemia (AML)**3—an aggressive cancer with a five-year survival rate of less than 30 % in adults.4
In recent years, several BRPF1b bromodomain inhibitors have been reported, including GSK68535 and NI-576 (Figure 1).
To discover new chemotypes, researchers have turned to in silico approaches such as high-throughput fragment docking7 and ligand-based screening.8 These efforts have been complemented by X-ray crystallography.
Despite these advances, no selective BRPF1b inhibitor has yet entered clinical trials. As a result, there is still strong interest in identifying and developing novel chemotypes that can selectively target this important epigenetic protein.
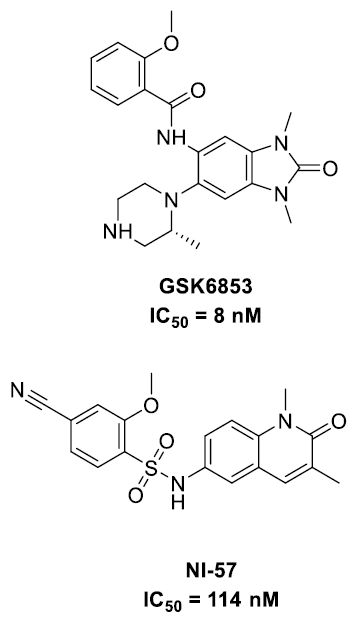
Figure 1. BRPF1b inhibitors. Image Credit: Concept Life Sciences
The BioPALS workflow
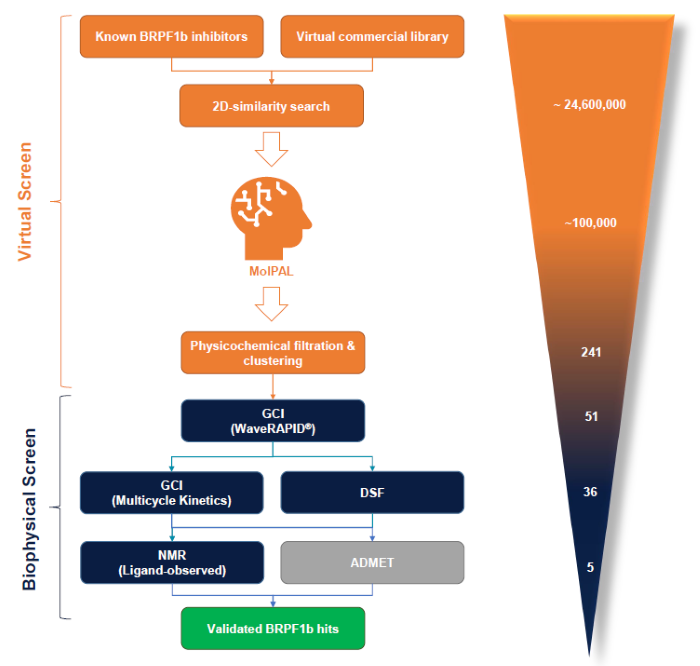
Figure 2. The BioPALS hit identification process is centred on the MolPAL algorithm and a GCI-driven biophysical hit confirmation workflow. In silico activities are displayed in orange, biophysical screening activities in blue and ADMET profiling in grey. The number of compounds processed though each individual stage for the BRPF1b bromodomain are shown on the right. Image Credit: Concept Life Sciences
AI-driven virtual screening via MolPAL
MolPAL9 is a structure-based virtual screening algorithm that uses simple molecular fingerprints to predict docking scores. It can efficiently identify top-scoring compounds while docking only a small fraction—typically around 1 %—of the full compound library.
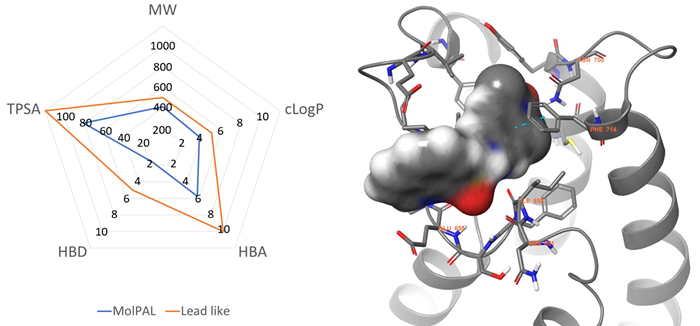
Figure 3. Left: Physicochemical profile of the MolPAL output (blue) compared to lead-like boundaries (orange). Right: The lowest energy docking pose was predicted by MolPAL for compound 5 (grey surface) within the BRPF1b active site (grey cartoon, PDB = 6EQK). Key residues are represented in grey sticks and amino acid sequence numbers with orange labels. Nitrogen, Oxygen, and Sulfur atoms are coloured blue, red, and yellow, respectively. π- π interactions are represented with dotted cyan lines. Image Credit: Concept Life Sciences
In the example presented here, an initial 2D-similarity search was used to narrow down 24.6 million commercial compounds to ~100,000 compounds. These compounds were then subjected to MolPAL (4 cycles) via Concept Life Sciences’ cloud computing platform.
A total of 51 diverse lead-like virtual hits were achieved via a subsequent chemoinformatic triage of the best virtual hits. Binding topologies for these hits were then predicted via MolPAL (Figure 3).
Primary in vitro confirmation via GCI
Grating-coupled interferometry (GCI) is a label-free biosensing technology capable of resolving extremely rapid binding kinetics with high precision. The WaveRAPID® (Repeated Analyte Pulses of Increasing Duration)10 method applies a single analyte concentration, delivered as a series of pulses with progressively increasing durations.
A WaveRAPID® GCI screen of the 51 virtual hits (100 µM) identified 36 primary binders to BRPF1b (Figure 4). Multicycle kinetics further confirmed that 20 of these compounds bound BRPF1b with KD values below 250 µM, corresponding to a 39 % hit rate. Binding kinetics were determined for all hits, alongside reference compounds GSK6853 and NI-57 (Table 1).
Selectivity was assessed using a BRPF1a counter-screen, which identified no binders—demonstrating that the newly identified hits are selective for BRPF1b over BRPF1a.
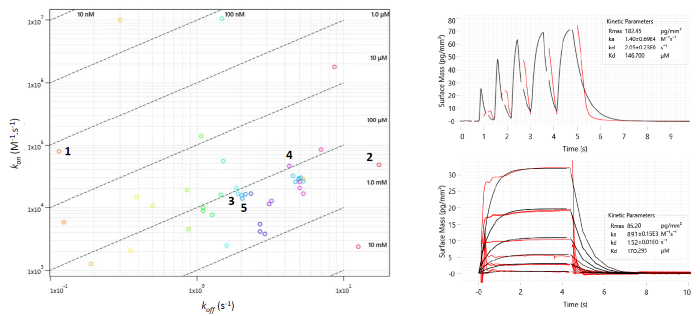
Figure 4. Left: Two-dimensional isoaffinity kinetic plot of association (kon) and dissociation (koff) rate constants from waveRAPID® screen of 51 virtual hits. The 36 primary hit binders are shown. Diagonal lines indicate equilibrium binding constants (KD). Each circle represents a binder and is coloured according to the dissociation rate. Top right: WaveRAPID® GCI sensorgram of compound 5 (100 μM) binding to BRPF1b. Bottom right: Multicycle kinetics GCI sensorgram of 5 binding to BRPF1b. Image Credit: Concept Life Sciences
Orthogonal hit confirmation via DSF
A total of 36 waveRAPID® hits were orthogonally confirmed via differential scanning fluorimetry (DSF) at 100 µM. This analysis saw 13 compounds induce a BRPF1b thermal shift (ΔTm) greater than ± 4 °C, signifying a notable interaction with the protein (Figure 5).
Four hits were shown to induce a thermal shift equal to or greater than NI-57 and GSK6853, while eight hits were found to exhibit a KD < 250 µM and a ΔTm > ±4 °C, including compounds 1, 2, 3, 4, and 5 (Table 1).
These results provided a prioritized list of BRPF1b hits, which were then confirmed in vitro via a combination of orthogonal techniques (GCI and DSF).
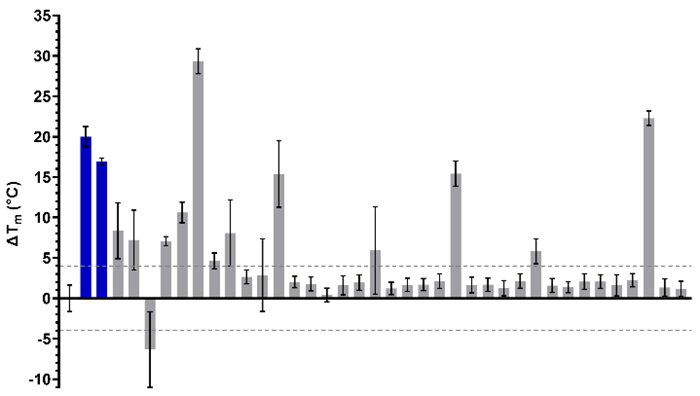
Figure 5. DSF characterization of the of 36 waveRAPID® primary hits (grey). GSK6853 and NI-57 are coloured in blue, and the > +4 °C and < -4 °C thresholds are indicated with grey dashed lines. Image Credit: Concept Life Sciences
Hit binding topology via ligand-observed NMR
Binding was further confirmed for four of the five prioritized hits via a range of ligand-observed NMR acquisition methods. These methods included 1D,11 STD,12 waterLOGSY,13 and CPMG14 in the presence and absence of BRPF1b protein.
It was possible to infer hit binding topologies for some of the ligands in accordance with specific shift changes following the addition of protein (Figure 6). This confirmed the docking poses previously predicted by MolPAL.
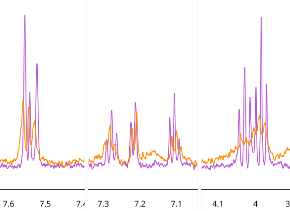
Figure 6. Sections of the 1D 1H NMR spectra illustrating peaks from compound 5 (400 μM) in the absence (pink) and presence (orange) of the BRPF1b Virtual commercial library bromodomain (26 μM). Image Credit: Concept Life Sciences
Selecting the best hits via biophysics and ADMET
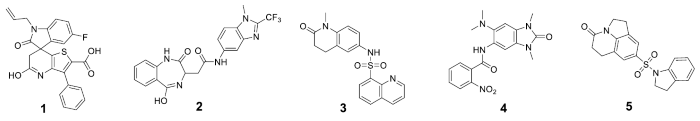
Image Credit: Concept Life Sciences
Table 1. Summary of biophysical data for GSK6853, NI-57 and five BRPF1b hits. cLogDs were determined using QikProp. KD, kon and koff were determined by GCI multicycle kinetics (N=2). ΔTm was determined by DSF (N=2). Ligand-observed NMR binding classification results from a consensus using 1D, STD, waterLOGSY and CPMG acquisition methods. n.d. = not determined. Source: Concept Life Sciences
| Entry |
MW |
cLogD |
KD |
kon |
koff |
ΔTm |
NMR binding |
| unit |
g.mol-1 |
- |
μM |
M.s-1 |
s-1 |
°C |
- |
| GSK6853 |
409 |
2.9 |
0.078 |
1.3 x 106 |
0.01 |
+ 19.9 |
n.d. |
| NI-57 |
383 |
1.5 |
0.086 |
1.9 x 107 |
0.16 |
+ 17.0 |
n.d. |
| 1 |
448 |
3.9 |
44 |
4.6 x 104 |
2.0 |
+ 7.2 |
Yes |
| 2 |
431 |
2.8 |
46 |
4.8 x 104 |
2.2 |
- 6.3 |
Yes |
| 3 |
367 |
2.1 |
61 |
3.5 x 104 |
2.0 |
+ 7.1 |
Insoluble |
| 4 |
369 |
2.9 |
128 |
3.0 x 104 |
2.1 |
+ 10.6 |
Yes |
| 5 |
354 |
2.1 |
186 |
8.4 x 103 |
1.6 |
+ 4.6 |
Yes |
Table 2. In vitro ADMET profile of five BRPF1b hits. CYP450 inhibition was measured as a pooled mixture of 1A2, 2C9 2C19, 2D6 and 3A4 isoforms. MPPB = mouse plasma protein binding, MLM = mouse liver microsomal stability, RLM = rat liver microsomal stability, HLM = human liver microsomal stability, HHeps = human hepatocyte stability, Fu = fraction unbound, n.d. = not determined. Source: Concept Life Sciences
| Entry |
MPPB
(Fu) |
MLM
(Clint) |
RLM
(Clint) |
HLM
(Clint) |
HHeps
(Clint) |
CYP450
inhib. (IC50) |
| unit |
- |
μL.min-1.mg-1 |
μL.min-1.mg-1 |
μL.min-1.mg-1 |
μL.min-1.106 cells |
μM |
| 1 |
0.18 |
17 |
41 |
< 5 |
< 3 |
All > 50 |
| 2 |
0.12 |
< 5 |
< 5 |
< 5 |
< 3 |
All > 10 |
| 3 |
0.23 |
20 |
89 |
10 |
4 |
n.d. |
| 4 |
0.12 |
6 |
< 5 |
< 5 |
< 3 |
n.d. |
| 5 |
0.05 |
157 |
216 |
94 |
67 |
n.d. |
Acknowledgments
Produced from materials originally authored Thomas Pesnot, Sandeep Pal, Zandile Nare, Vincenzo A. Rao, Ian Morrison, Andrew Scott, Matilda J. Bingham, and Muhammad Jan from Concept Life Sciences Integrated Discovery & Development Services Ltd.; Brian O. Smith from the School of Molecular Biosciences, College of Medical Veterinary and Life Sciences, University of Glasgow; and Edward A. Fitzgerald from Malvern Panalytical.
References and further reading
- You, L., et al. (2015). Deficiency of the Chromatin Regulator Brpf1 Causes Abnormal Brain Development. Journal of Biological Chemistry, 290(11), pp.7114–7129. https://doi.org/10.1074/jbc.m114.635250.
- Cheng, C.L.-H., et al. (2021). Bromodomain-containing protein BRPF1 is a therapeutic target for liver cancer. Communications Biology, 4(1). https://doi.org/10.1038/s42003-021-02405-6.
- Shima, H., et al. (2013). Bromodomain-PHD finger protein 1 is critical for leukemogenesis associated with MOZ–TIF2 fusion. International Journal of Hematology, (online) 99(1), pp.21–31. https://doi.org/10.1007/s12185-013-1466-x.
- Cancer Research UK (2016). Acute myeloid leukaemia (AML) | Cancer Research UK. (online) Cancerresearchuk.org. Available at: https://www.cancerresearchuk.org/about-cancer/acute-myeloid-leukaemia-aml.
- Bamborough, P., et al. (2016). GSK6853, a Chemical Probe for Inhibition of the BRPF1 Bromodomain. ACS Medicinal Chemistry Letters, (online) 7(6), pp.552–557. https://doi.org/10.1021/acsmedchemlett.6b00092.
- Igoe, N., et al. (2017). Design of a Chemical Probe for the Bromodomain and Plant Homeodomain Finger-Containing (BRPF) Family of Proteins. Journal of Medicinal Chemistry, 60(16), pp.6998–7011. https://doi.org/10.1021/acs.jmedchem.7b00611.
- Zhu, J. and Caflisch, A. (2016). Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions. Journal of Medicinal Chemistry, 59(11), pp.5555–5561. https://doi.org/10.1021/acs.jmedchem.6b00215.
- Zhu, J., Zhou, C. and Caflisch, A. (2018). Structure-based discovery of selective BRPF1 bromodomain inhibitors. European Journal of Medicinal Chemistry, 155, pp.337–352. https://doi.org/10.1016/j.ejmech.2018.05.037.
- Graff, D.E., Shakhnovich, E.I. and Coley, C.W. (2021). Accelerating high-throughput virtual screening through molecular pool-based active learning. Chemical Science, 12(22), pp.7866–7881. https://doi.org/10.1039/d0sc06805e.
- Kartal, Ö., et al. (2021). waveRAPID—A Robust Assay for High-Throughput Kinetic Screens with the Creoptix WAVEsystem. SLAS DISCOVERY: Advancing the Science of Drug Discovery, 26(8), pp.995–1003. https://doi.org/10.1177/24725552211013827.
- Hajduk, P.J., Olejniczak, E.T. and Fesik, S.W. (1997). One-Dimensional Relaxation- and Diffusion-Edited NMR Methods for Screening Compounds That Bind to Macromolecules. Journal of the American Chemical Society, 119(50), pp.12257–12261. https://doi.org/10.1021/ja9715962.
About Concept Life Sciences
Concept Life Sciences is a leading contract research organisation (CRO) serving the global life sciences industry. For over 25 years, the company, and its heritage companies, have provided consultative and collaborative drug discovery and development services. Our approach, supported by passionate scientists and world-leading capabilities, enables clients to overcome complex scientific challenges across a broad range of therapeutic areas, improving program success rates. The company has successfully helped 29 candidates advance to the clinic.
The company offers sophisticated translational biology services coupled with exceptional end-to-end chemistry capabilities across all modalities, including small molecules, biologics, peptides and cell & gene therapies, with the ability to seamlessly integrate capabilities and provide bespoke solutions to address client needs.
Collectively, the company’s high-quality services and commitment to customer service across the drug development pathway enhance efficiency in drug discovery, helping clients advance their drugs to clinic in as little as 32 months, well ahead of the industry average of 60 months.
Driven by a passion for science, Concept Life Sciences has around 230 employees, with around 70% holding PhDs. The company operates from state-of-the-art UK facilities, headquartered near Manchester, with additional operations in Edinburgh, Dundee, and Sandwich. The headquarters is one of the UK’s largest medicinal chemistry CRO sites with key discovery services all under one roof.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.