Coronavirus disease 2019 (COVID-19) caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) presents a significant threat to global public health. As of today, it has caused more than 40 million confirmed cases and over 1.12 million deaths across the world. Since there are no drugs or vaccines for COVID-19 yet, current treatment strategies mainly focus on supportive care, symptom management, and isolation of infected people.
SARS-CoV-2 is a positive-sense, single-stranded, enveloped RNA virus. It has a spike (S) protein that mediates the virus's entry into the host cell by binding to the angiotensin-converting enzyme 2 (ACE2) in the host cell membrane. This significant function of the S protein makes it a significant target in the efforts to develop drugs and vaccines for COVID-19.
Recently, researchers from the Lehigh University, Pennsylvania, USA; Seoul National University, Republic of Korea; Korean Institute of Science and Technology Information, Republic of Korea; and University of Cambridge, UK reported a study in which they analyzed the structure and dynamics of the spike protein using multiple molecular dynamic simulations. Their study is published on the preprint server bioRxiv.*
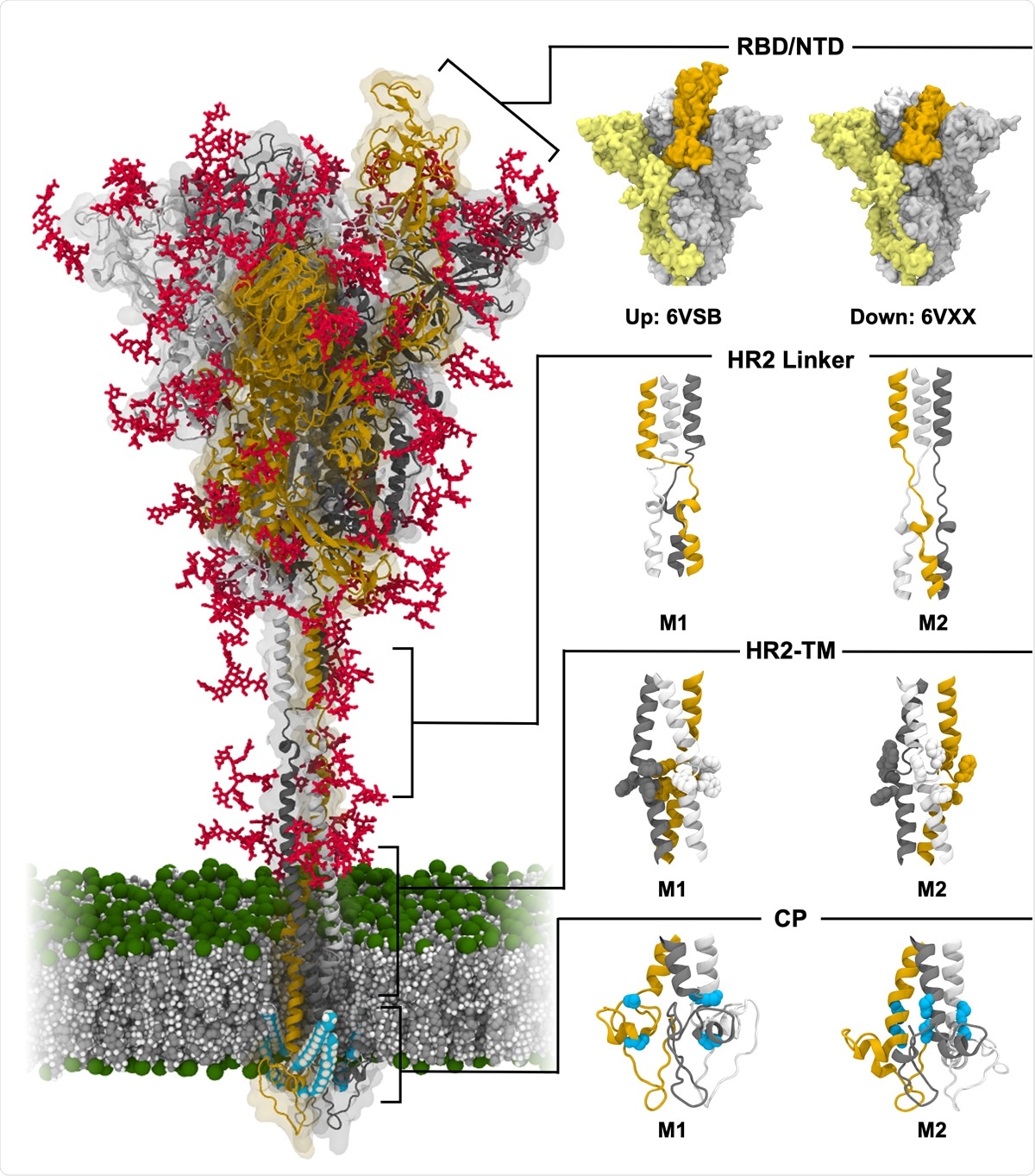
Model structure of fully-glycosylated full-length SARS-CoV-2 S protein in a viral membrane. A model structure of SARS-CoV-2 S protein is shown on the left panel. Two models for RBD/NTD, HR2 linker, HR2-TM, and CP are enlarged on the right panel. The three individual chains of S protein are colored in yellow, gray, and white, respectively, while glycans are represented as red sticks. The palmitoylation sites of S protein are highlighted in cyan. The phosphate, carbon, and hydrogen atoms of the viral membrane are colored in green, gray, and white, respectively. For clarity, water molecules and ions are omitted. All illustrations were created using Visual Molecular Dynamics (VMD)30.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Overall shape and orientation of the S protein are determined by the flexible stalk
The authors had previously developed fully glycosylated full-length models of SARS-CoV-2 S protein in a viral membrane with different templates for the stalk region and both open and closed conformations receptor-binding domain (RBD). In this study, they performed multiple µs-long, all-atom molecular dynamic simulations to gain a more in-depth insight into the structure and dynamics of the spike protein and the functions of glycan.
The simulations revealed that the highly flexible stalk determines the S protein's overall shape and orientation on the membrane surface with 2 independent joints. More importantly, the S protein models in the simulations allow prediction of possible configurations of S protein-ACE2-B0 AT1 complex. The simulations also helped identify glycans attached to different glycosylation sites that stabilize RBD's open and closed conformations by creating a high-energy barrier between the open-closed transition.
"Our MD simulations reveal the overall shape of S protein and its orientation on the membrane surface are determined by a highly flexible stalk composed of two independent joints."
According to the team, their simulations show that the S protein's most probable orientations are capable of ACE2 binding. They identified multiple glycans that helped stabilize the RBD and showed that the exposure of antibody epitopes could be analyzed using a detailed study of the antibody-glycan clash. Although accessible surface area (ASA) analysis is the usual method used, it overestimates the impact of glycan shielding and does not take into account the possible detailed interactions between the antibody and glycan.
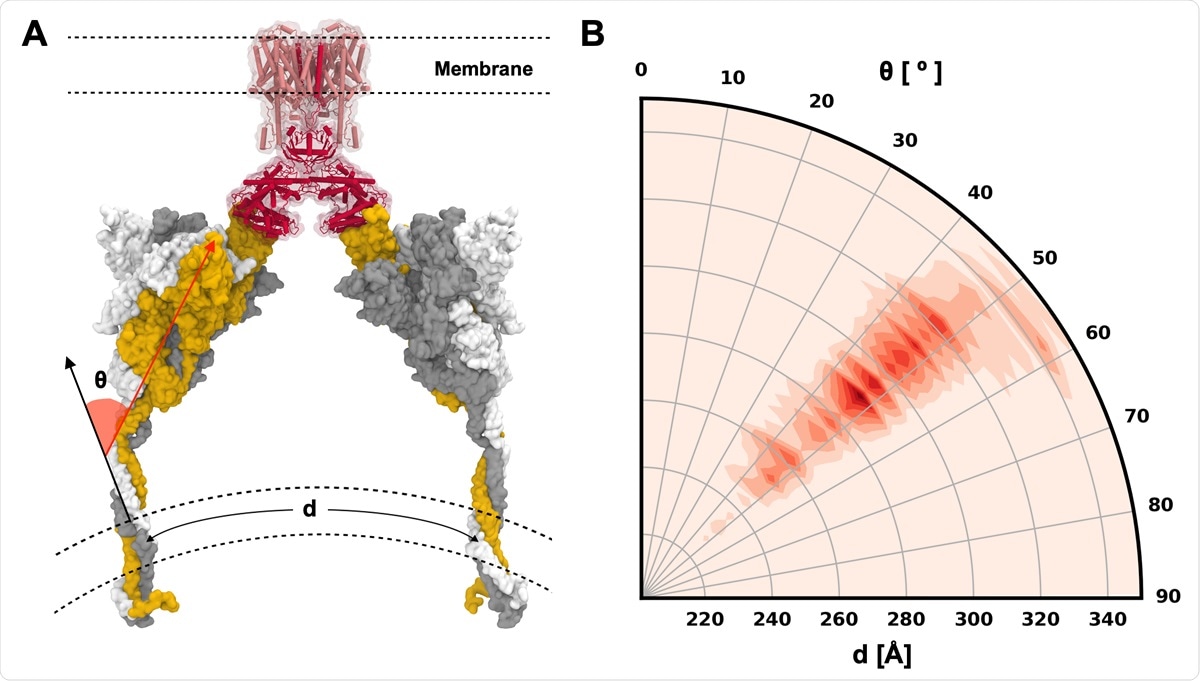
S protein configurations competent for ACE2 binding. (A) Illustrative snapshot of S protein-ACE2-B0AT1 complex. Three individual chains of S protein are colored in yellow, gray, and white, and ACE2 and B0AT1 are represented as red and pink, respectively. (B) Distribution of tilting angle (θ) as a function of the arc length (d) between the centers of mass (COMs) of TM domains.
Simulations offer crucial insights into S protein structure and dynamics
The researchers presented all-atom MD simulations of fully glycosylated and full-length S protein in a viral bilayer in this work. Also, it generated multiple µs-long trajectories for the RBD in open and closed states. They also performed many µs-long simulations of non-glycosylated S protein head-only systems. They showed that upon removal of glycans, the open-state RBD becomes unstable and transition to the close state at the early stage. The team showed that the impact of glycan shielding is overestimated in the ASA analysis by aligning RBD-antibody complex structures to the simulation trajectories.
The researchers say that their results enable a better understanding of the functional roles of glycans that can not only shield for immune evasion but also offer stability to the trimer and help with the transition of RBD open and close conformations. Their simulations also provided insights into key structural roles of the highly flexible stalk conformations in spike protein binding to ACE2. Overall, the study's observations offer deeper insights into the structure and dynamics of the SARS-CoV-2 S protein, and these insights can potentially guide the design of antiviral therapeutics for COVID-19.
"Our results provide deeper insight into functional roles of glycans that provide not only shielding for immune evasion but also contribute to the trimer stability and transition of RBD open and close states. "

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Structure, Dynamics, Receptor Binding, and Antibody Binding of Fully-glycosylated Full-length SARS-CoV-2 Spike Protein in a Viral Membrane Yeol Kyo Choi, Yiwei Cao, Martin Frank, Hyeonuk Woo, Sang-Jun Park, Min Sun Yeom, Tristan I. Croll, Chaok Seok, Wonpil Im bioRxiv 2020.10.18.343715; doi: https://doi.org/10.1101/2020.10.18.343715, https://www.biorxiv.org/content/10.1101/2020.10.18.343715v1
- Peer reviewed and published scientific report.
Choi, Yeol Kyo, Yiwei Cao, Martin Frank, Hyeonuk Woo, Sang-Jun Park, Min Sun Yeom, Tristan I. Croll, Chaok Seok, and Wonpil Im. 2021. “Structure, Dynamics, Receptor Binding, and Antibody Binding of the Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein in a Viral Membrane.” Journal of Chemical Theory and Computation 17 (4): 2479–87. https://doi.org/10.1021/acs.jctc.0c01144. https://pubs.acs.org/doi/10.1021/acs.jctc.0c01144.