The COVID-19 pandemic caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has crippled global healthcare systems and triggered an economic crisis in many countries. The pandemic is not showing signs of a slowdown even in 2021 with a new and more infectious variant of the virus wreaking havoc in the UK and many other parts of the world.
The mortality rates of COVID-19 vary across the globe and range from 0.5% to 13%, according to Johns Hopkins University estimates. COVID-19 has a variable clinical presentation with the majority of patients experiencing very mild disease, many asymptomatic, and a few patients experiencing severe, life-threatening disease requiring hospitalization and intensive care. The most powerful determinant of COVID-19 disease severity is age, with individuals over 70 years more likely to develop severe COVID-19. Host factors contributing to severe disease may include variations in the immune response owing to immunological history and genetic determinants.
The patient’s immune system status is a significant factor in COVID-19 infection survival. More and more studies are focusing on immunological determinants of severe disease in order to develop better biomarkers for therapeutic purposes. The mechanism of the insurgence of immunity is at the center of the development of both drugs and vaccines to treat or prevent SARS-CoV-2 infection.
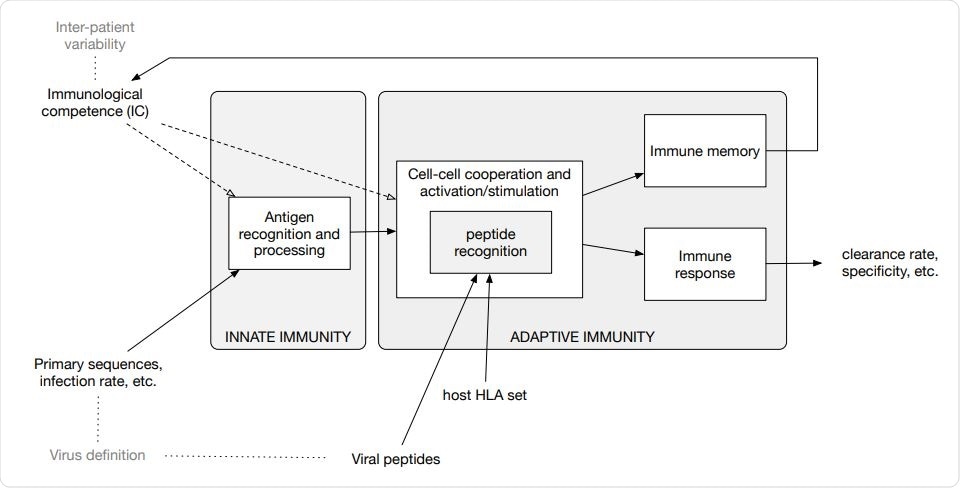
Diagram of the in-silico model components and accepted input. The model embodies functions to calculate the clonal affinity to precomputed viral peptides of the selected pathogen (defined by its primary sequence) with respect to a specific HLA set. The population-dynamics of the elicited lymphocytes clones, resulting from the infection by the SARS-CoV-2, provides a varying degree of efficiency of the immune response which, as it turns out, correlates with the parameters defining both the immunological competence (IC) of the virtual host and the virus definition.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Simulating the SARS-CoV-2 immune response to analyze impact of viral load and age on disease severity
A team of researchers from Italy, India, and the United Kingdom recently simulated the SARS-CoV-2 immune response and analyzed the impact of the viral load, ACE2 receptor affinity, and age during the course of the disease in an artificially infected population. The study has been published on the preprint server bioRxiv*.
This study addresses fundamental aspects of infection management in terms of describing the insurgence and the span of immunity in COVID-19. The in silico model developed by the researchers focuses on this aspect at both the individual and population levels.
The team used a stochastic agent-based immune simulation platform to build a virtual cohort of infected individuals with age-dependent, varying degrees of immune response. They used a parameter setting to reproduce known inter-patient variability and general epidemiological statistics.
Antibody titers post day 25 of infection is a prognostic factor for determining COVID-19 outcome
The researchers made several clinical observations in silico and identified critical factors in the statistical evolution of COVID-19 infection. They found evidence for the significance of the humoral response over the cytotoxic response and also observed that the measurement of antibody titers post day 25 of infection is a prognostic factor for ascertaining the clinical outcome of the infection.
The team’s modeling framework uses SARS-CoV-2 infection to illustrate the actionable effectiveness of simulating the immune response at both individual and population levels. The model developed by the team can explain and interpret observed infection patterns and makes verifiable temporal predictions.
“The results of a large number of simulations that we call virtual or in-silico cohort, demonstrates that the great variability observed in the real pandemic can be the mere result of such diversity in both viral and human characteristics.”
Findings highlight the significance of model predictions
According to the authors, this study proposes quantitatively that the high variability observed in clinical outcomes of COVID-19 in real life can be due to the subtle differences in viral load and immune competence of the population.
To summarize, this work shows the power of model predictions, identifies the clinical endpoints ideal for computational modeling of the immune response to COVID-19, and defines the resolution and amount of data needed to empower such models for translational medicine purposes. The researchers also demonstrate how computational modeling of immune response offers key insights to discuss hypotheses and develop new experiments.
“Despite the extraordinary complexity of the immune system dynamics, the progress of simulation platforms suggests that a more intense interaction between clinicians and researchers in the computational model could bring these models to the desired quality for deployment in the medical field.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
From infection to immunity: understanding the response to SARS-CoV2 through in-silico modeling Filippo Castiglione, Debashrito Deb, Anurag P. Srivastava, Pietro Liò, Arcangelo Liso bioRxiv 2020.12.20.423670; doi: https://doi.org/10.1101/2020.12.20.423670, https://www.biorxiv.org/content/10.1101/2020.12.20.423670v1
- Peer reviewed and published scientific report.
Castiglione, Filippo, Debashrito Deb, Anurag P. Srivastava, Pietro Liò, and Arcangelo Liso. 2021. “From Infection to Immunity: Understanding the Response to SARS-CoV2 through In-Silico Modeling.” Frontiers in Immunology 12 (September). https://doi.org/10.3389/fimmu.2021.646972. https://www.frontiersin.org/articles/10.3389/fimmu.2021.646972/full.