Digital PCR (dPCR) is an exceptionally accurate and sensitive method ideally suited for identifying copy number variations (CNVs), also referred to as copy number alterations (CNAs).

The Digital LightCycler® dPCR System. Image Credit: Roche Sequencing and Life Science
CNVs/CNAs encompass genomic regions where sequences have undergone deletions or amplifications, and they are associated with various health conditions, including cancer and numerous neurodevelopmental disorders.
The ability to detect even subtle changes in copy number variation is crucial for identifying risk factors and devising effective treatments.
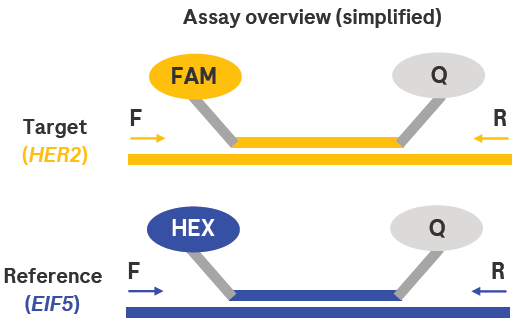
Figure 1. Simplified assay overview. Here, two targets are detected at the same time (multiplexing). Target-specific primers (arrows) extend during amplification, dislodging labeled probes from target sequences. The probes release a dye-specific signal (FAM or HEX) that is detected by the instrument. The Qs represent quencher molecules, which prevent dye activation until the probe is released. Image Credit: Roche Sequencing and Life Science
Multiplexing allows the simultaneous detection of two targets. During amplification, target-specific primers (represented by arrows) extend and displace labeled probes from the target sequences. These probes release a dye-specific signal (FAM or HEX) detected by the instrument. The Qs symbolize quencher molecules, which prevent dye activation until the probe is released.
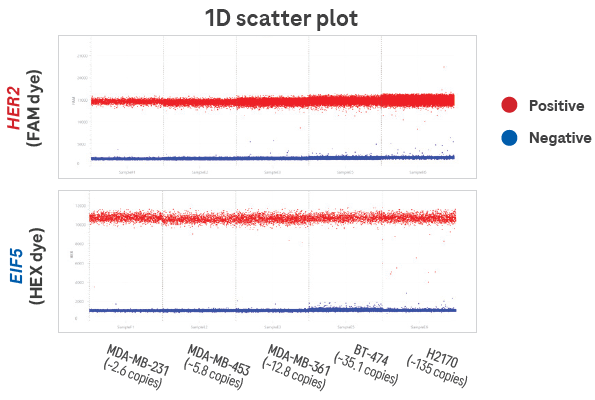
Figure 2. Qualitative 1D scatter plot showing distinct clustering and minimal rain. The results show increased HER2 signal relative to EIF5 in proportion to copy number. Each section of the chart represents one sample, and each dot represents one partition. The distinct clustering and minimal rain demonstrate optimal assay performance and high signal-to-noise ratio. 8 ng of cell line DNA was included per filling reaction on the 100,000-well High-Resolution Plate (cell lines are labeled above with expected HER2 copy number). Image Credit: Roche Sequencing and Life Science
The results demonstrate an increased HER2 signal relative to EIF5 in proportion to the copy number. Each section of the chart represents a sample, with each dot indicating a partition.
The distinct clustering and minimal rain demonstrate optimal assay performance and a high signal-to-noise ratio. For the filling reaction, 8 ng of cell line DNA was included per partition, utilizing the 100,000-well High-Resolution Plate (cell lines labeled above with expected HER2 copy number).
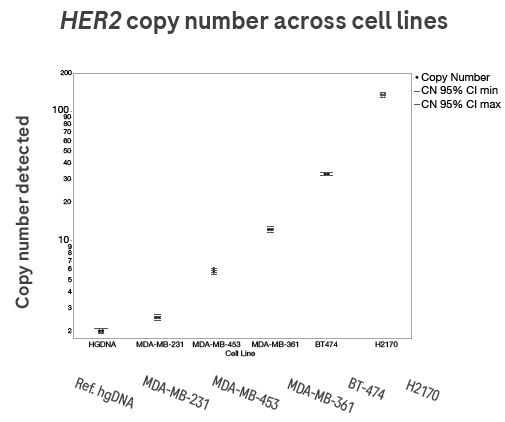
Figure 3. Quantitative analysis of dPCR results across a wide range of copy numbers. Here, wild-type hgDNA (copy number = 2) is used as a reference. For this analysis, the High-Resolution Plate (with 100,000 partitions) was used, as it enables clear discrimination over a wide range of HER2 copy numbers, ~2 to ~135. Image Credit: Roche Sequencing and Life Science
In this case, wild-type hgDNA (copy number = 2) serves as a reference. The analysis employs the High-Resolution Plate (with 100,000 partitions) to enable precise discrimination across a wide range of HER2 copy numbers, approximately 2 to 135.
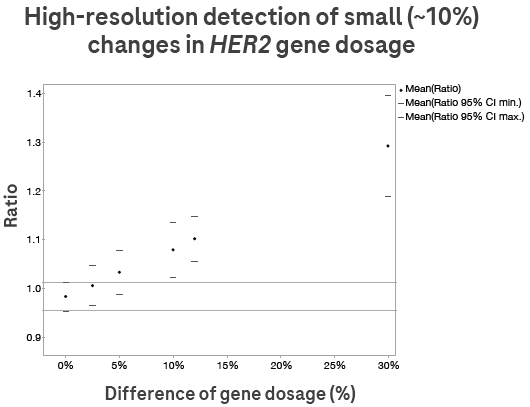
Figure 4. Digital LightCycler® dPCR System detects copy number changes as small as 10%. Samples with varied copy numbers were created by spiking human gDNA with MD-MB-231 cell line DNA in specific ratios. See Figure 6 for an overview of how copy number is calculated. Image Credit: Roche Sequencing and Life Science
Samples with varying copy numbers were created by introducing MD-MB-231 cell line DNA into human gDNA at specific ratios. Refer to Figure 6 for an overview of the copy number calculation process.
Digital LightCycler® workflow
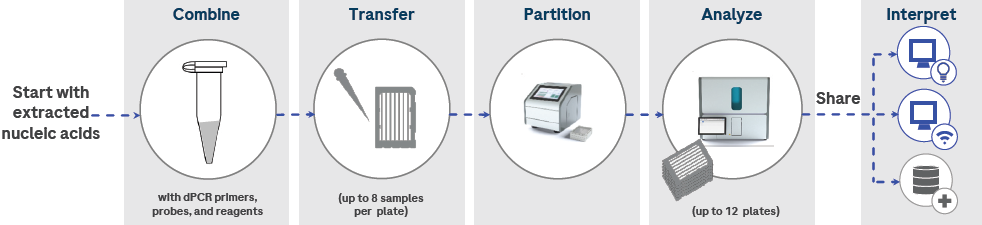
Figure 5. An overview of the Digital LightCycler® dPCR System workflow. Extracted nucleic acids are combined with PCR reagents, including primers and probes. The reaction mixture is transferred to partitioning plates and partitioned on the Partitioning Engine. Plates are transferred to the Analyzer and processed with the appropriate cycles. The collected data is then analyzed with Digital LightCycler® software and web applications, and can be shared via LIMS. Image Credit: Roche Sequencing and Life Science
Nucleic acids obtained from extraction are combined with PCR reagents, including primers and probes. The reaction mixture is then transferred to partitioning plates and partitioned using the Partitioning Engine.
Following that, the plates are transferred to the Analyzer for processing with the appropriate cycles. The collected data is subsequently analyzed using Digital LightCycler® software and web applications, facilitating convenient data sharing through LIMS.
How copy number is calculated
After signal detection on the Analyzer, Poisson distribution calculations are employed to determine the total number of signal incidents. Subsequently, a target/reference ratio is utilized to determine the copy number.
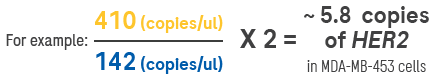
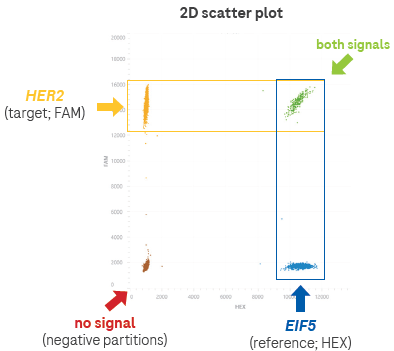
Figure 6. An example of how copy number is calculated using a 2-plex assay. In the 2D scatter plot (one way of visualizing the data from the Analyzer), each dot represents a detection event. The analysis software calculates the absolute number of incidents of each signal, and then a ratio is calculated to provide the copy number of the target. Image Credit: Roche Sequencing and Life Science
In the 2D scatter plot (one of the visualization methods for Analyzer data), each dot represents a detection event. The analysis software calculates the absolute number of signal incidents, which are then used to calculate the ratio and determine the copy number of the target.
About Roche Sequencing and Life Science

Roche Sequencing & Life Science is part of Roche Diagnostics, which, along with Roche Pharmaceuticals, plays an important role in modern healthcare. Roche Diagnostics’ broad range of innovative diagnostic tests and systems play a pivotal role in the groundbreaking area of integrated healthcare solutions and cover the early detection, targeted screening, evaluation and monitoring of disease. Roche Diagnostics is active in all market segments, from scientific research and clinical laboratory systems to patient self-monitoring.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.