Mapping autophagy in action
Autophagy, the process by which cells remove damaged organelles and pathogens, plays a vital role in maintaining cellular health. Yet its role during viral infection has remained elusive. Autophagy is crucial for immunity and the removal of viruses from the cell's cytoplasm, a process known as virophagy.1 When a viral infection occurs, the innate immune system can trigger autophagy to break down the invading viruses.
Herpesviruses have evolved strategies to sometimes evade autophagic degradation, such as by producing particular proteins that inhibit autophagy.
Two human herpesviruses that can lead to cancer are Epstein-Barr virus (EBV) and Kaposi sarcoma-associated herpesvirus (KSHV), both of which are part of the gamma-herpesvirus sub-family.
Kaposi sarcoma-associated herpesvirus (KSHV) is linked to several conditions, including Kaposi’s sarcoma (KS), B-cell primary effusion lymphoma (PEL), multicentric Castleman’s disease (MCD), and osteosarcoma. KSHV is present globally but is less widespread than other herpesviruses, with a unique regional distribution. Worldwide, KSHV seroprevalence is generally below 10 %, though it is higher among men who have sex with men. In sub-Saharan Africa and Mediterranean regions, seroprevalence can vary from 20 % to 80 % across both genders. The virus is primarily transmitted through saliva.2
Currently, the autophagy process during KSHV entry into host cells remains unclear. Additionally, there is no curative treatment or vaccine for KSHV infection, highlighting the importance of understanding early immune responses to KSHV entry. This understanding could potentially lead to new targets for antiviral therapies.
Researchers at the University of Zurich3 visualized the rapid activation of autophagy within minutes of KSHV entering host cells. The autophagy marker, cytosolic microtubule-associated protein light chain 3 (LC3), was shown to co-localize with viral capsid proteins, signaling that the cell’s defense machinery was engaging incoming viruses.
Through correlative confocal microscopy using the Thermo Scientific Helios 5 Hydra UX DualBeam (focused ion beam-scanning electron microscope - FIB-SEM) and reconstructed in Thermo Scientific Amira Software, scientists produced high-resolution 3D models revealing that KSHV particles reside within amphisomes, fusion vesicles formed between autophagosomes and endosomes. These structures indicate that cells actively target virus-damaged compartments for degradation.
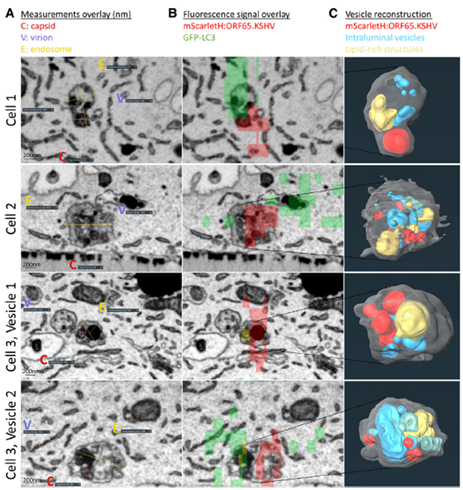
Figure 1. Ultrastructural images of KSHV-containing vesicles and their three-dimensional reconstructions reveal amphisomal structures. Image Credit: Figure reproduced from Schmidt et al. under CC-BY 4.0.
Galectin-8 and NDP52: Key players in cellular defense
The study identified galectin-8 and the receptor NDP52 as critical mediators of the autophagic response to KSHV. When the virus disrupts endosomal membranes, galectin-8 detects the damage and recruits NDP52, which in turn mobilizes autophagy machinery to degrade the infected vesicles.
Through quantitative co-localization analysis, the researchers were able to visualize and analyze molecular interactions, confirming that the galectin-8-NDP52 partnership is central to early antiviral defense.
Advancing discovery through 3D imaging and analysis
The integrative imaging workflow, linking confocal microscopy, electron microscopy, and image segmentation in Amira Software, was key to visualizing the sequence of molecular events that drive viral restriction. Amira Software enabled the team to:
- Correlate fluorescence and electron microscopy data for precise structural insight
- Quantify morphometric data of viral particles and vesicles
- Visualize dynamic cell processes, including endosomal damage and autophagosome recruitment.
These capabilities confirmed that selective autophagy serves as a rapid, effective antiviral defense, intercepting KSHV before infection is established.
Shaping the future of infectious disease research
The discovery that selective autophagy restricts KSHV infection through the recruitment of galectin-8 and NDP52 marks a significant step forward in understanding how cells defend themselves against viral invasion. With Amira Software, scientists can explore these complex biological processes in remarkable 3D detail, transforming visualization into a powerful tool for discovery.
Learn more about visualization and analysis of viral infection mechanisms at thermofisher.com/amira.
Watch on-demand how to maximize your cell biology images with Amira Software
References
- Lussignol, M. and Esclatine, A. (2017). Herpesvirus and Autophagy: ‘All Right, Everybody Be Cool, This Is a Robbery!’ Viruses, 9(12), p.372. DOI: 10.3390/v9120372. https://www.mdpi.com/1999-4915/9/12/372
- Cesarman, E., Damania, B., Krown, S.E., Martin, J., Bower, M., and Whitby, D. (2019). Kaposi sarcoma. Nature Reviews Disease Primers, [online] 5(1), pp.1–21. DOI: 10.1038/s41572-019-0060-9. https://www.nature.com/articles/s41572-019-0060-9.
- Schmidt, K.W., et al. (2024). Selective autophagy impedes KSHV entry after recruiting the membrane damage sensor galectin-8 to virus-containing endosomes. Cell Reports, 43(12), pp.115019–115019. DOI: 10.1016/j.celrep.2024.115019. https://www.sciencedirect.com/science/article/pii/S2211124724013706.

Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.