Innovative imaging techniques unveil neural network coordination in planaria regeneration
Planarians are flatworms that have a simple central nervous system (CNS) but an extraordinary ability to regenerate neural tissue. Their CNS is divided into distinct molecular and functional domains defined by specific neural gene expressions 1.
Remarkably, planarians can regenerate functional brains from small body fragments, thanks to pluripotent stem cells known as neoblasts, which can differentiate into various cell types, including neurons. As a result, planarians have become a model organism for studying neural regeneration 2-3.
However, exploring the complex coordination of neural networks in planarian growth and regeneration has always been a challenge, primarily due to the difficulties in capturing high-resolution, three-dimensional images of their central nervous system (CNS) within a practical timeframe.
To overcome these obstacles, researchers from Zhejiang and Westlake University and Tottori University 4 used advanced imaging techniques combining tissue expansion with tiling light-sheet microscopy, enabling nanoscale resolution and significant linear expansion.
Central to this success was Thermo Scientific's Amira Software, which served as the primary environment for image processing and analysis, particularly for segmentation, quantification, and 3D visualization.
Automated 3D cell segmentation to study neuron and muscle fiber distribution in planarians
By implementing an automated 3D cell segmentation process, researchers mapped neurons and muscle fibers at the single-cell level in over 400 wild-type planarians, both during their normal state and throughout regeneration.
They revealed the complex distribution of neurons across the planarian nervous system, including the brain, ventral nerve cords, optic nerves, and pharyngeal nerve complex. This innovative approach promises to shed new light on the intricate processes underlying planarian biology.
By creating recipes in Amira Software, researchers were able to quantify neurons and nuclei and to collect volumetric parameters such as length and volume.
The recipe solution in Amira Software provides a powerful tool that significantly enhances the efficiency and consistency of image analysis workflows. By allowing users to create, save, and reuse analysis recipes, it ensures that complex image processing tasks can be replicated across different samples with minimal effort.
Analyzing neural and muscle fiber differences using Amira Software
Using Amira Software capabilities in image analysis and processing, researchers identified differences between a control sample and one where β-catenin, which is crucial for maintaining structural integrity and functional interactions between neurons and muscle fibers, was interfered with (as shown in the figure below).
Specifically, they observed that neural structures have a clear and organized pattern in the control, highlighting the distinct pathways and connections within the sample. In contrast, in β-catenin RNAi planarian, muscle fibers show a more complex and cobweb-like structure, indicating a different or more intricate arrangement of neural connections.
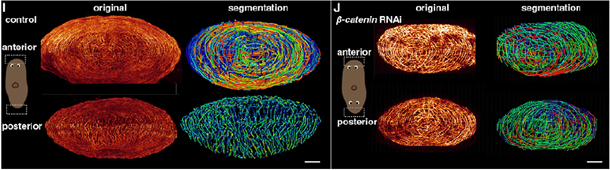
Segmentation of anterior and posterior muscle fibers in the control and in β-catenin-1 RNAi planarian. Image Credit: Figure from Lu et al. Under CC BY 4.0 license
To support open science and reproducibility, the research team released all Amira Software segmentation and quantification recipes via Zenodo.
From 3D images to 3D understanding
Lu et al.’s work clearly demonstrates how Amira Software can bridge advanced microscopy and data science, delivering quantitative, reproducible workflows that illuminate how cells, neurons, and tissues organize in three dimensions.
From robust nuclei counting to fiber tracing and 3D rendering, Amira Software enabled the researchers to uncover structural principles of regeneration previously unseen in living systems. This workflow now serves as a model for future imaging studies seeking reproducibility, scalability, and cross-modality integration.
Discover how Amira can enhance your 3D imaging workflows, from segmentation to analysis to visualization at thermofisher.com/amira.
References
- Cebria, F., et al. (2002). Dissecting planarian central nervous system regeneration by the expression of neural-specific genes. Development, Growth and Differentiation, 44(2), pp.135–146. DOI: 10.1046/j.1440-169x.2002.00629.x. https://onlinelibrary.wiley.com/doi/10.1046/j.1440-169x.2002.00629.x.
- Umesono, Y. and Agata, K. (2009). Evolution and regeneration of the planarian central nervous system. Development, Growth & Differentiation, 51(3), pp.185–195. DOI: 10.1111/j.1440-169x.2009.01099.x. https://onlinelibrary.wiley.com/doi/10.1111/j.1440-169X.2009.01099.x.
- Cebrià, F. (2007). Regenerating the central nervous system: how easy for planarians! Development Genes and Evolution, 217(11-12), pp.733–748. DOI: 10.1007/s00427-007-0188-6. https://link.springer.com/article/10.1007/s00427-007-0188-6.
- Lu, J., et al. (2025). 3D reconstruction of neuronal allometry and neuromuscular projections in asexual planarians using expansion tiling light sheet microscopy. eLife, (online) 13, p.RP101103. DOI: 10.7554/eLife.101103. https://elifesciences.org/articles/101103.

Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.