Drug metabolism is a central factor in drug discovery and development, impacting safety, pharmacokinetics, and pharmacodynamics. Research into drugs’ in vitro metabolism in human and animal tissues supports the identification of major metabolism pathways, also referred to as ‘soft spots’.1
The Mass-MetaSite software uses a number of algorithms to detect the peaks corresponding to metabolites in mass spectra acquired from incubated samples. This process includes background subtraction, isotope pattern analysis, noise suppression, retention time analysis, and mass shift analysis, based on cyp, non-cyp, and other uncommon reactions.
Potential metabolites are scored based on the number of matches between its fragments and the parent compound. The Mass-MetaSite software generates site of metabolism (SoM) predictions that can be used to distinguish between potential regioisometric metabolites that exhibit identical fragmentation patterns and mass shifts.
These tools streamline the process of metabolite identification. This article presents an efficient workflow for identifying soft spots using EAD, a novel orthogonal fragmentation technique. This workflow is completed using the ZenoTOF 7600 system, with the Mass-MetaSite software used to predict sites of metabolism.
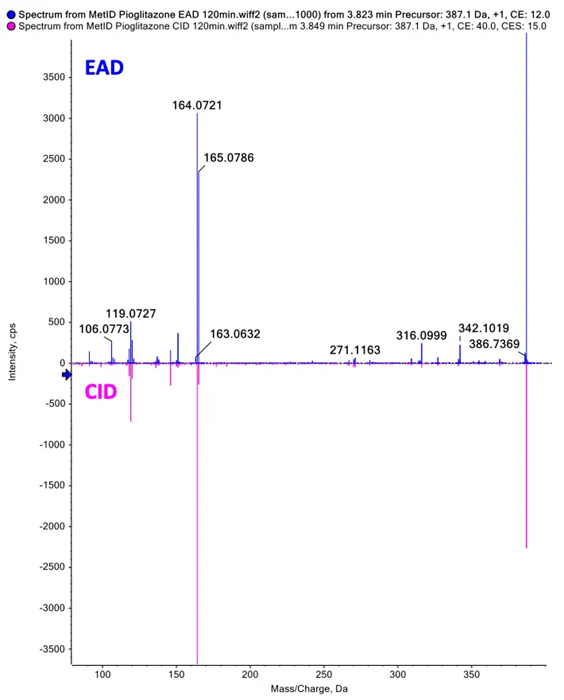
Figure 1. EAD provided rich MS/MS spectra for confident identification of pioglitazone phase 1 metabolites. EAD (top) and CID (bottom) spectra for a metabolite showing aliphatic hydroxylation and alcoholic oxidation at a retention time of 3.77 minutes. Image Credit: SCIEX
Key features of the ZenoTOF 7600 system for metabolite identification
- Confident identification: More fragments are acquired with EAD, allowing users to confidently identify the site of metabolism for phase 1 metabolites. EAD spectra have been shown to be more informative for metabolite identification than collision-induced dissociation (CID) spectra.
- Rapid characterization and identification: The ZenoTOF 7600 system has been designed to offer rapid and efficient software-aided characterization and identification of drug metabolites from hepatocyte incubations.
- Detection of low-level metabolites: The Zeno trap’s enhanced MS/ MS sensitivity enables the identification of low-level metabolites present in drug metabolism studies.
- Streamlined data acquisition and processing workflow: The instrument’s quick, easy-to-use workflow from acquisition to analysis enables the development of confident structure-metabolic stability relationships for drugs under investigation.
Methods
- Sample preparation: Pioglitazone was incubated in human hepatocytes at 37 °C, at a starting concentration of 5 µM. Samples were removed from incubation at 0-, 30-, 60-, 90-, and 120-minute intervals and quenched with acetonitrile.
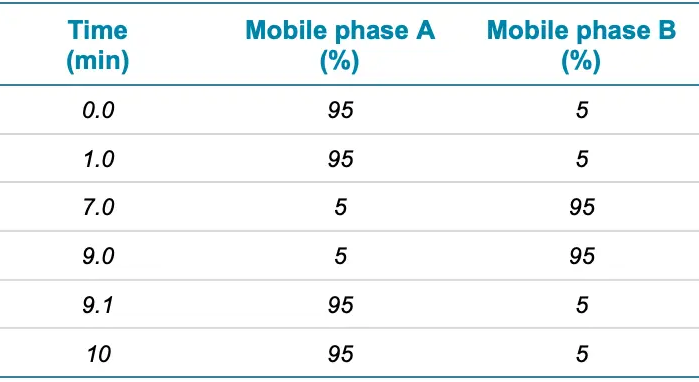
- Chromatography: A Phenomenex Kinetex Polar C18 column (2.1 x 100 mm, 2.6 µm, 100 Å) was used to perform separation, at a column temperature of 40 °C. Mobile phase A was 0.1 % (v/v) formic acid in water, while mobile phase B was 0.1 % (v/v) formic acid in methanol. A 5 µL injection was selected for analysis. Table 1 summarizes the chromatographic gradient conditions employed.
Table 1. Chromatographic gradient. Source: SCIEX
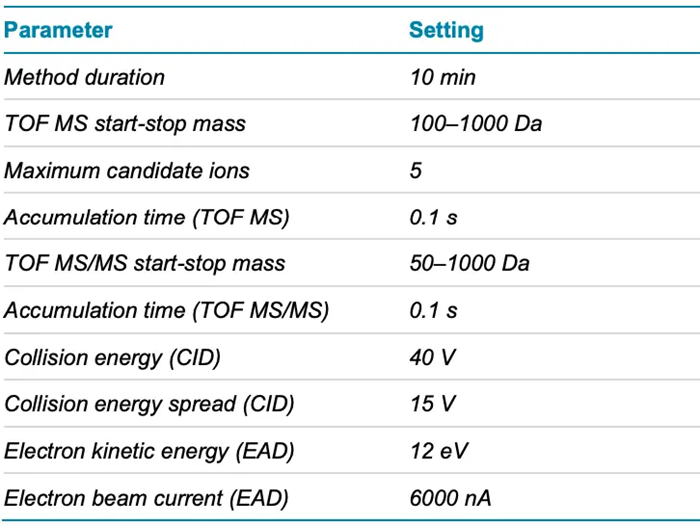
- Mass spectrometry: Samples were analyzed using the ZenoTOF 7600 system’s data-dependent acquisition (DDA) method, which included Zeno CID DDA and Zeno EAD DDA. Table 2 summarizes the source and gas conditions used, while Table 3 shows the method conditions.
Table 2. Source and gas conditions. Source: SCIEX
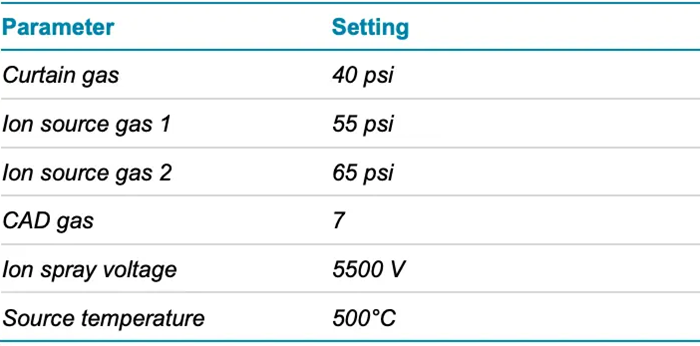
Table 3. Zeno DDA parameters. Source: SCIEX
- Data processing: Version 3.0 of the SCIEX OS software was used for data acquisition, with the Mass-MetaSite software employed for predicting biotransformation sites using Zeno CID DDA and Zeno EAD DDA data.4-9
Using positional information acquired via EAD to identify the site of metabolism
Following the acquisition of data, results from both EAD and CID analyses were compared to assess the positional information provided by each technique. Three peaks at retention times of 3.53, 3.77, and 4.13 minutes were identified as hydroxy pioglitazone in the 120-minute incubation sample, using Zeno CID DDA and Zeno EAD DDA data.
Zeno EAD data for the hydroxy pioglitazone peak at 3.53 minutes yielded a total of 10 product ion matches, prompting the software to label this peak hydroxy pioglitazone (M-VII).
Zeno CID data prompted the software to predict two possibilities for hydroxylation, which included hydroxy pioglitazone (M-II) and hydroxy pioglitazone (M-IV). These two structures were predicted with equal likelihoods and seven product ion matches per structure (Figure 2).
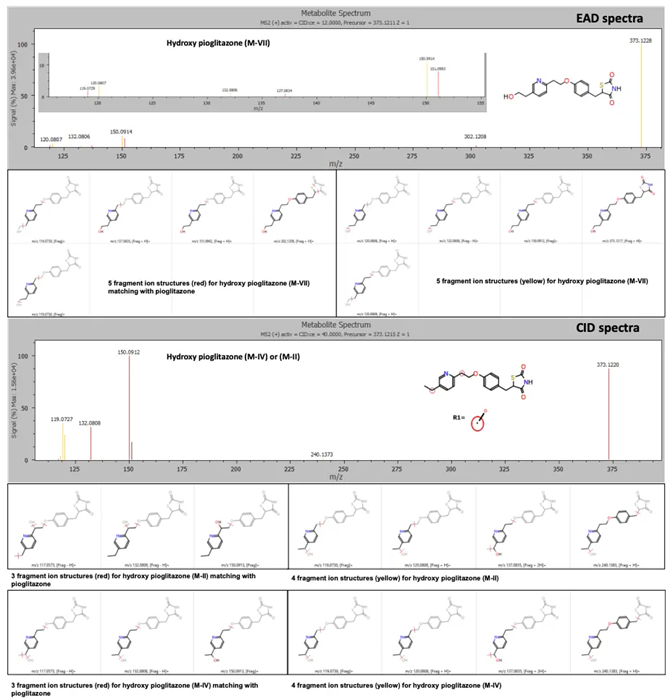
Figure 2. EAD and CID spectra for the hydroxy pioglitazone metabolite at retention time 3.53 minutes with fragment ion matches for structures predicted by Mass-MetaSite software. Product ion matches with pioglitazone are displayed in red and metabolite-specific matches are indicated in yellow. Image Credit: SCIEX
The hydroxy pioglitazone peak at a retention time of 3.77 minutes was scored highest for hydroxy pioglitazone (M-II) based on a total of six product ion matches. The hydroxy pioglitazone (M-II) was ranked second with a total of six fragment matches from CID data.
EAD data highlighted six possible structures for the hydroxyl metabolite, with hydroxy pioglitazone (M-IV) ranked first with the highest score and a total of seven product ion matches (Figure 3).
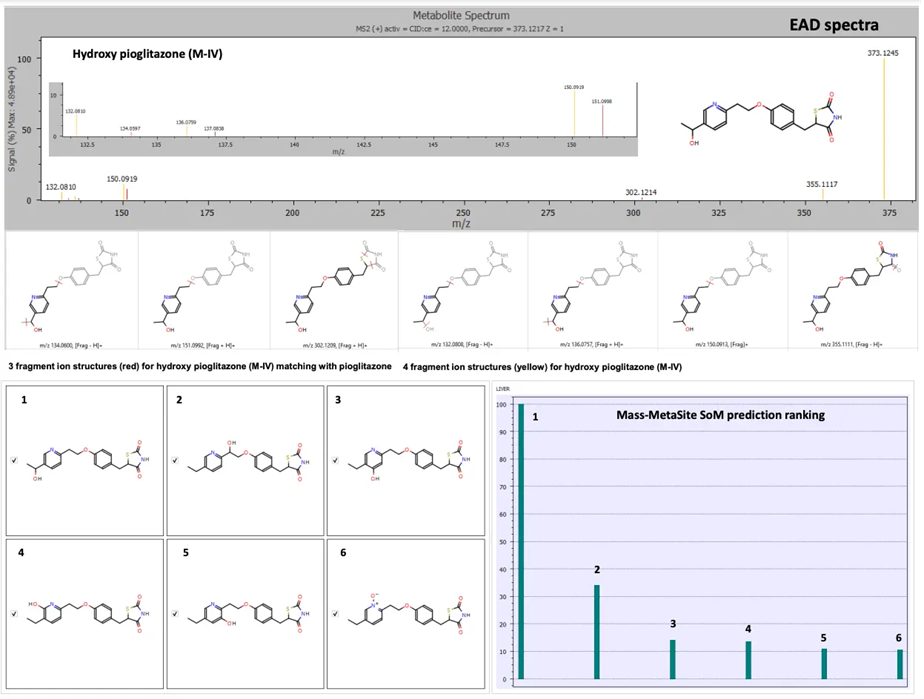
Figure 3. EAD spectra for the hydroxy pioglitazone metabolite at retention time 3.77 minutes with fragment ion matches for structures predicted by Mass-MetaSite software and SoM prioritization ranking. Product ion matches with pioglitazone are displayed in red and metabolite-specific matches are indicated in yellow. The SoM prioritization ranking indicated the highest probability match for hydroxy pioglitazone (M-IV). Image Credit: SCIEX
The hydroxy pioglitazone peak at a retention time of 4.13 minutes was labeled as either potential amide hydrolysis and dehydrogenation, based on six product ion matches, or hydroxy pioglitazone (M-II), with a lower likelihood based on five product ion matches from the CID data.
EAD data revealed a higher likelihood that the peak actually corresponded to hydroxy pioglitazone (M-II), based on a total of eight product ion matches (Figure 4).
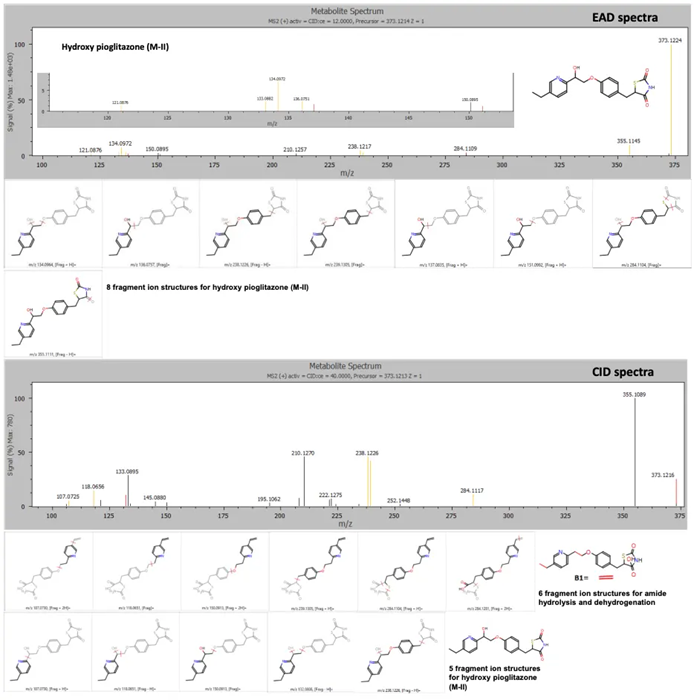
Figure 4. EAD and CID spectra for the hydroxy pioglitazone metabolite at retention time 4.13 minutes with fragment ion matches for structures predicted by Mass-MetaSite software. Product ion matches with pioglitazone are displayed in red and metabolite-specific matches are indicated in yellow. Image Credit: SCIEX
Both EAD and CID spectra were used to label a metabolite with aliphatic hydroxylation and alcoholic oxidation at a retention time of 3.78 minutes. The rich MS/ MS spectra acquired via EAD facilitated a more confident structure assignment, with EAD data yielding a total of eight fragment matches, versus the three fragment matches yielded by CID data (Figure 5).3-9
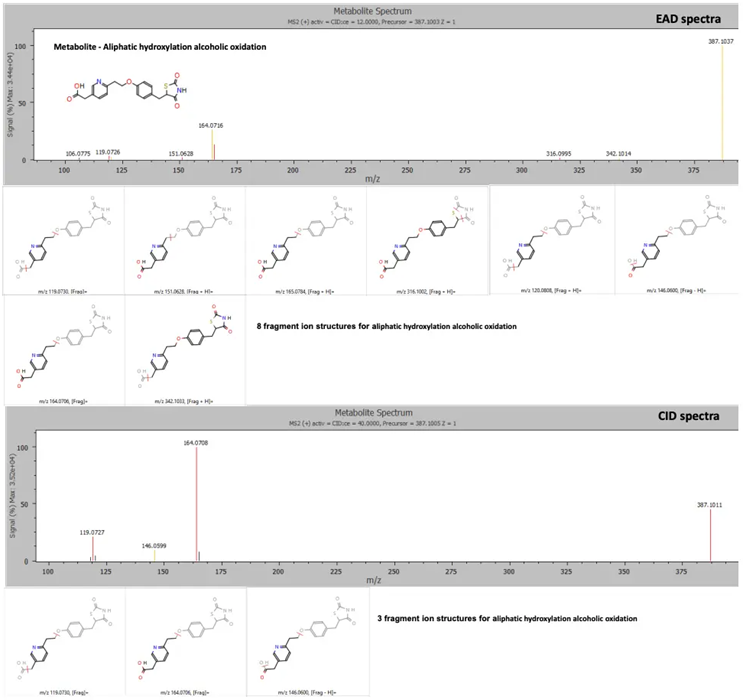
Figure 5. EAD and CID spectra for a metabolite showing aliphatic hydroxylation and alcoholic oxidation with fragment ion matches for structures predicted by Mass-MetaSite software. Product ion matches with pioglitazone are displayed in red and metabolite-specific matches are indicated in yellow. Image Credit: SCIEX
It was possible to confirm all three pioglitazone hydroxy metabolites (M-VII, M-IV, and M-II) by matching their respective retention times to those of standard solution injections.
The ZenoTOF 7600 system exhibited excellent mass accuracy for the workflow, with the full range of metabolites and fragments identified with <two ppm error. This accuracy allowed the confident identification of phase one metabolites present in an in vitro metabolism study of pioglitazone.
It was also possible to easily and confidently identify phase one metabolites with the MS/ MS coverage offered by Zeno EAD on the ZenoTOF 7600 system.
Conclusion
EAD’s ability to provide more information-rich product ion spectra supported the rapid software-aided identification and characterization of phase 1 metabolites from hepatocyte incubations of pioglitazone on the ZenoTOF 7600 system.
The workflow presented in this article can be easily adapted for in vivo metabolism studies, enabling the detection of low-level metabolites due to the Zeno trap’s improved sensitivity.
This streamlined data acquisition and processing workflow facilitates expedited data reduction, enabling the development of confident structure-metabolism stability relationships for pharmaceutical drugs.
References and further reading
- Zhang, Z. and Tang, W. (2018). Drug metabolism in drug discovery and development. Acta Pharmaceutica Sinica B, (online) 8(5), pp.721–732. DOI: 10.1016/j.apsb.2018.04.003. https://www.sciencedirect.com/science/article/pii/S2211383517306767?via%3Dihub.
- Causon, J. et al. (2021). Orthogonal fragmentation mechanism enables new levels of metabolite characterization, SCIEX. https://sciex.com/content/dam/SCIEX/tech-notes/pharma/discovery/ruo-mkt-02-13348-a/Darunavir-MetID_ZenoTOF-7600_RUO-MKT-02-13348-A.pdf.
- Bonn, B., et al. (2010). Enhanced metabolite identification with MSE and a semi-automated software for structural elucidation. Rapid Communications in Mass Spectrometry, 24(21), pp.3127–3138. DOI: 10.1002/rcm.4753. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/rcm.4753.
- Zamora, I., et al. (2013). High-throughput, computer assisted, specific MetID. A revolution for drug discovery. Drug Discovery Today: Technologies, 10(1), pp.e199–e205. DOI: 10.1016/j.ddtec.2012.10.015. https://www.sciencedirect.com/science/article/abs/pii/S1740674912000911?via%3Dihub.
- Strano-Rossi, S., et al. (2014). Metabolism of JWH-015, JWH-098, JWH-251, and JWH-307 in silico and in vitro: a pilot study for the detection of unknown synthetic cannabinoids metabolites. Analytical and Bioanalytical Chemistry, 406(15), pp.3621–3636. DOI: 10.1007/s00216-014-7793-9. https://link.springer.com/article/10.1007/s00216-014-7793-9.
- Zelesky, V., et al. (2013). Software automation tools for increased throughput metabolic soft-spot identification in early drug discovery. Bioanalysis, (online) 5(10), pp.1165–79. DOI: 10.4155/bio.13.89. https://www.tandfonline.com/doi/full/10.4155/bio.13.89.
- Li, A.C., et al. (2012). Update on hydrocodone metabolites in rats and dogs aided with a semi-automatic software for metabolite identification Mass-MetaSite. Xenobiotica, 43(4), pp.390–398. DOI: 10.3109/00498254.2012.715697. https://www.tandfonline.com/doi/full/10.3109/00498254.2012.715697.
- Brink, A., et al. (2014). Post‐acquisition analysis of untargeted accurate mass quadrupole time‐of‐flight MSE data for multiple collision‐induced neutral losses and fragment ions of glutathione conjugates. Rapid Communications in Mass Spectrometry, 28(24), pp.2695–2703. DOI: 10.1002/rcm.7062. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/rcm.7062.
- Ge, S., Tu, Y. and Hu, M. (2016). Challenges and Opportunities with Predicting In Vivo Phase II Metabolism via Glucuronidation From In Vitro Data. Current Pharmacology Reports, 2(6), pp.326–338. DOI: 10.1007/s40495-016-0076-8. https://link.springer.com/article/10.1007/s40495-016-0076-8.
Acknowledgments
Produced from materials originally authored by Rahul Baghla and Eshani Nandita from AB Sciex LLC.
About SCIEX
SCIEX's mission is to deliver solutions for the precision detection and quantification of molecules, empowering its customers to protect and advance the wellness and safety of all.
SCIEX has led the field of mass spectrometry for 50 years. From the moment it launched the first ever commercially successful triple quad in 1981, SCIEX has developed groundbreaking technologies and solutions that influence life-changing research and outcomes.
Today, as part of the Danaher family of global life science and technology innovators, it continues to pioneer robust solutions in mass spectrometry and capillary electrophoresis. But SCIEX doesn’t just develop products. It is what the company does together with its customers that sets it apart. That’s why thousands of life science experts around the world choose SCIEX to get the answers they can trust to better inform critical decisions. Decisions that positively impact lives.
SCIEX stands proudly by its tagline: The Power of Precision.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.