With the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) across most of the world, scientists are working strenuously to find effective therapies or vaccines. Most strategies being worked on at present are targeting viral entry into the host cells, mainly through the interaction of the SARS-CoV-2 S protein with the angiotensin-converting enzyme 2 (ACE2) receptor.
The entry of the virus is a complex process with multiple steps, each working in symphony on different stages of the S protein. The protein has two subunits, S1 and S2, and the S1 has a receptor-binding domain (RBD). This loop-dominant RBD consisting of 273 amino acids, is only part of the whole S protein, extending from residue 319 to 591. This region is key to the protein-receptor binding.
In order to develop drugs that act on the binding process, it may be fruitful to examine RBD residues that are separate from the receptor-binding motif (RBM) but constrain this region. Therefore, in a new study published on the preprint server bioRxiv* in June 2020, the researchers from India and the United States split off the RBD from the main protein in order to critically examine it in detail.
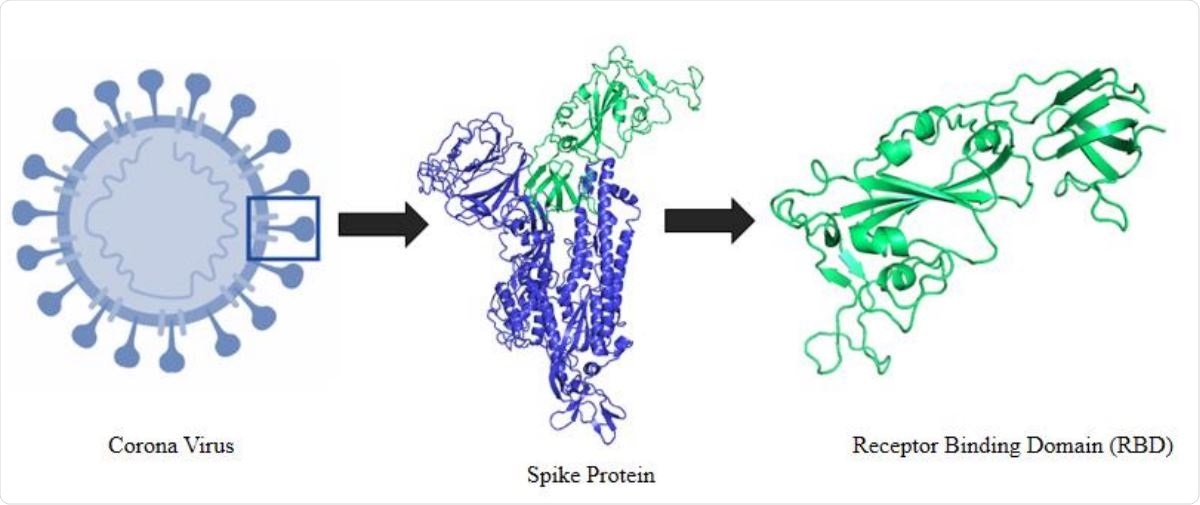
Schematic representation of the selection logistics of the protein and protein segment. It shows an animated figure of the 2019 novel coronavirus. Segment presented in green cartoon refers to the receptor binding domain (RBD) of the spike protein.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Co-Evolutionary And Sequence Analysis
The investigators used two levels of study techniques to understand the evolution of the RBD as well as its structural basis. It is known that the RBD undergoes processing and then docks into the ACE2 receptor on the host cell, in a binding cleft. It then sets the stage for the process of viral entry.
The first or evolutionary study aimed to understand the sequence space of the RBD by recreating it so that the closest sequences in other strains and viruses could be seen. This helped to understand the evolutionary constraints, which is a crucial step to understand how far a protein can evolve. This is required to plan a counter-strategy that will discourage evolution.
This showed that many flexible regions of disorder are present, mostly as RBD loops, with the largest at the C-terminal end and a central region about 60 residues long.
The investigators used a co-evolutionary analysis so that positionally interdependent regions can be identified. This, in turn, provides an ongoing picture of interrelated residues. Such interdependence is required to stabilize the protein structure and predict the functionalities of the protein.
On the other hand, sequence conservation analysis provides a snapshot of the residue positions that are so important that zero variation is allowed. The use of both these analytic pathways was aimed at understanding how the RBD sequence space was constrained and how it was changing.
Important Structure Blocks Identified
The results showed that the highest interdependence and co-evolutionary potential was between region 121 and 180 of the 273-residue RBD. The mutational profile was also generated, which could allow the scientists to visualize the tolerance for substitution mutations at each site in this region.
They attempted to find all the natural mutations interacting with the host ACE2 receptor and to identify their impact by statistical methods using energy computations from the deep mutational scan performed earlier. They found that there is a high tolerance for positional residue substitution.
Finally, they matched critical co-evolutionary residues with their mutational tolerance. Highly conserved regions had a lower tolerance and vice versa
The RBD structure was also used to divide the S protein into 30 sub-structures, using interacting pairs, so that clustering of residues by pair-wise interactions as well as by regions with high local variability could be identified. Such regions were discovered to be near the residues of the RBD binding cleft, from the docking data.
SARS-CoV-2 viruses binding to ACE-2 receptors on a human cell, the initial stage of COVID-19 infection, conceptual 3D illustration credit: Kateryna Kon / Shutterstock
How The Protein Contributes To Infection
The importance of this particular segment, called SB6, in adding local conformational flexibility to the protein, so as to bind efficiently with the ACE2 receptor, was suggested from this finding, which is more specific than the earlier reports ascribing importance to the loops in general. In the words of the researchers, “SB6, with its balanced abundance of conserved and co-evolving residues, can be hypothesized to be a critical cluster, which is further validated by its constituent residues, which impose a significant impact on the residue interacting with ACE2 receptor.”
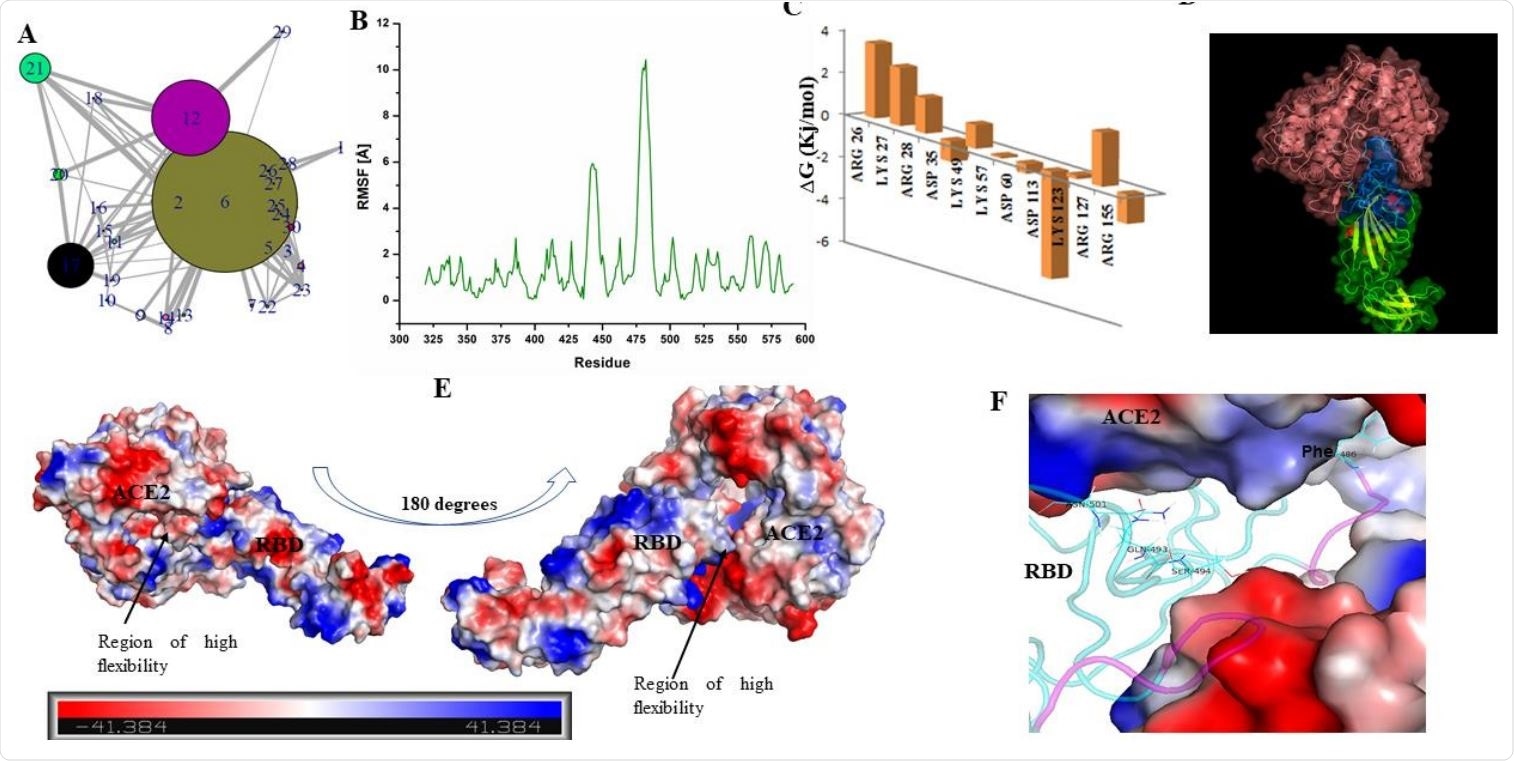
The structure of RBD has loop dominant region, which plays a critical role in ACE2 receptor recognition and associated conformational dynamics. (A) Structure Network plot of spike RBD. (B) Root mean square fluctuations (in Angstrom) for the spike RBD as retrieved from Monte Carlo simulation. (C) Bar plots showing the effects of destabilizing mutations. (D) Docked complex of Spike RBD-ACE2 is shown in surface representation with cartoons representing the protein secondary structures. ACE2 receptor is shown in pink while RBD is shown as blue and green surfaces. Blue surface refers to the region in the RBD which participates in ACE2 receptor binding. (E) Docked complex is shown with surface charge representations. Region of high flexibility as observed from MC simulation is marked in the docked complex. Scale bar in the bottom refers to the gradient of surface charge as observed in the docked complex. (F) RBD-ACE2 interaction pocket (ACE2 is shown as surface and RBD in cartoon) is shown with purple coloured loops indicating substructure with high flexibility (as predicted from RMSF).
This region also overlaps with the stretches that show high coevolution signal in SB17 and SB 21, demonstrating its importance in viral evolution. This is very disorganized, and consists mostly of loops, making it very flexible and capable of receptor recognition.
The unified approach showed that destabilization of the RBD would have a deep effect on both the evolution and the structure of the S protein, as well as preventing its receptor recognition functionality. This shows it to be a promising therapeutic target.
The researchers comment, “We strongly hypothesize that SB17 is an extremely critical component of the spike RBD structure, which could be an important therapeutic target. Unlike the other reports specifically pointing the residues associated with ACE2 receptor recognition, we provided an extended picture of regions in the protein structure, which, if targeted, could potentially inhibit virus propagation and block the probable mutational escape routes, thereby functioning as an anti-evolution strategy.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources