Researchers in the United States have identified cardiopulmonary processes that may partly explain why some individuals with coronavirus disease 2019 (COVID-19) develop respiratory failure.
The SNPs were filtered using a relaxed p-value to increase the amount of potential genomic information generated. The p-value is a measure used to express the statistical significance of evidence.
This enabled the identification of a unique set of genes that could then be analyzed to determine biological correlates that may be linked to pulmonary or cardiac symptoms.
A pre-print version of the article is available in the server medRxiv*, while the article undergoes peer review.
The interplay between severe COVID-19 and comorbidities is not well understood
Since the first cases of COVID-19 were identified in Wuhan, China, late last year, the causative agent –severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – has spread at an unprecedented rate causing worldwide public health and economic crisis.
While the majority of infected individuals are asymptomatic or only experience mild symptoms, around 5% develop severe lung injury and acute respiratory distress syndrome, which can be fatal.
The most commonly reported health conditions associated with such severe outcomes include hypertension, diabetes, cardiovascular disease, and chronic respiratory infection. However, the molecular mechanisms underlying severe COVID-19 and their interplay with these comorbidities are not well understood.
To investigate the main genetic variants associated with respiratory failure, an international consortium group carried out a GWAS of participants with severe acute respiratory failure due to COVID-19 at seven hospitals in Italy and Spain.
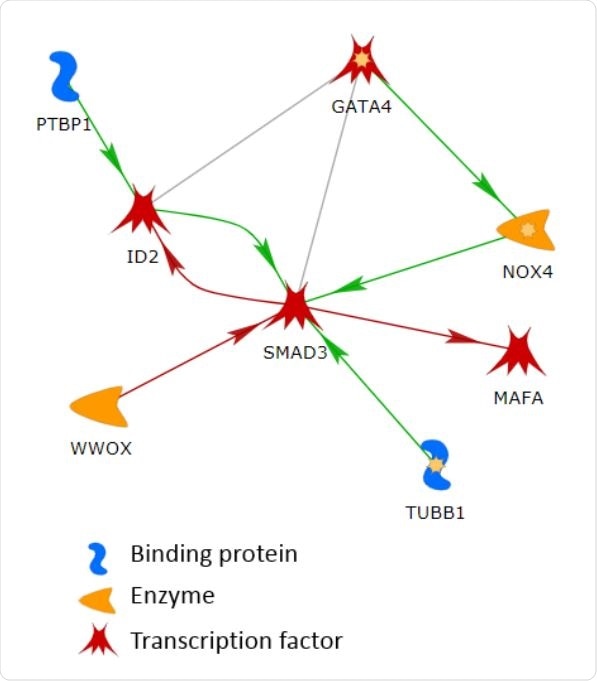
The largest connected network in the list of 144 genes. The line colors indicate the activation (green), inhibition (red), and unspecified (grey) effects.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
What did the current study involve?
Jung Hun Oh (Memorial Sloan Kettering Cancer Center) and colleagues assessed the GWAS summary statistics available for 835 COVID-19 patients with respiratory failure and 1,255 controls.
The statistics included information on individual loci and p-values for 8,582,968 SNPs that had been submitted to the European Bioinformatics Institute.
The researchers downloaded the summary statistics and, to enrich biological discovery, employed a relaxed p-value of 5×10-5 as a filtering threshold on the statistics.
This “likely enabled the inclusion of much potential genomic information in the analysis and the identification of plausible biological correlates associated with pulmonary or cardiac symptoms,” they write.
The SNPs generated were then mapped to the nearest genes, and 144 unique genes were identified.
The top-ranked biological correlates relevant to respiratory failure
Gene ontology and protein-protein interaction (PPI) analyses performed on these genes revealed that the top-ranked biological correlates relevant to respiratory failure were wound healing, epithelial structure maintenance, muscle system processes, and cardiac-related processes.
The PPI analysis found that the largest connected network among the 144 genes consisted of 8 genes, including GATA4, ID2, MAFA, NOX4, PTBP1, SMAD3, TUBB1, and WWOX.
Next, a PubMed literature search was conducted to see whether there were any previous reports on the biological associations between these genes and respiratory or cardiac symptoms.
The search revealed that 7 of the 8 genes had previously been related to both pulmonary and cardiac diseases.
For example, one study reported that GATA4 functions as a transcription factor in normal pulmonary development, while another study identified it as a candidate gene relevant to congenital heart disease.
One study reported that in the case of chronic obstructive pulmonary disorder, increased expression of NOX4 and TGF-β correlated with increased volume of airway smooth muscle mass and epithelial cells. Another study found that the upregulation of NOX4 in the heart promoted cardiac remodeling, indicating a potential role in reducing the severity of heart failure.
What are the implications of the study?
The researchers say the evidence suggests that in severe cases of COVID-19, lung disease is likely to be associated with cardiovascular risk.
“Further research is warranted to identify the common biological processes between lung and heart diseases and the interplay between them,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources