Researchers in the United States have conducted a study suggesting that the host response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) within nasal mucosa is a key determinant of clinical trajectory in coronavirus disease 2019 (COVID-19).
The team’s analysis of nasopharyngeal samples from a cohort of individuals with COVID-19 revealed both protective and detrimental host responses to SARS-CoV-2 within the nasal epithelium and identified direct targets of infection.
Overall, the findings point to the early failure of antiviral responses within nasal epithelial cells as a predictor of progression to severe disease.
“Together, our work demonstrates that many of the factors associated with the clinical trajectory following SARS-CoV-2 infection may stem from initial host-viral encounters in the nasopharyngeal epithelium,” writes Jose Ordovas-Montanes from the Broad Institute of MIT and Harvard in Cambridge, Massachusetts and colleagues.
The team says the findings also suggest that therapeutic interventions targeting the nasopharynx during the early stage of infection may prevent severe disease.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
The potential significance of the impact on airway epithelium
Infection with SARS-CoV-2 can cause severe disease characterized by lower respiratory symptoms, including pneumonia and acute respiratory distress syndrome.
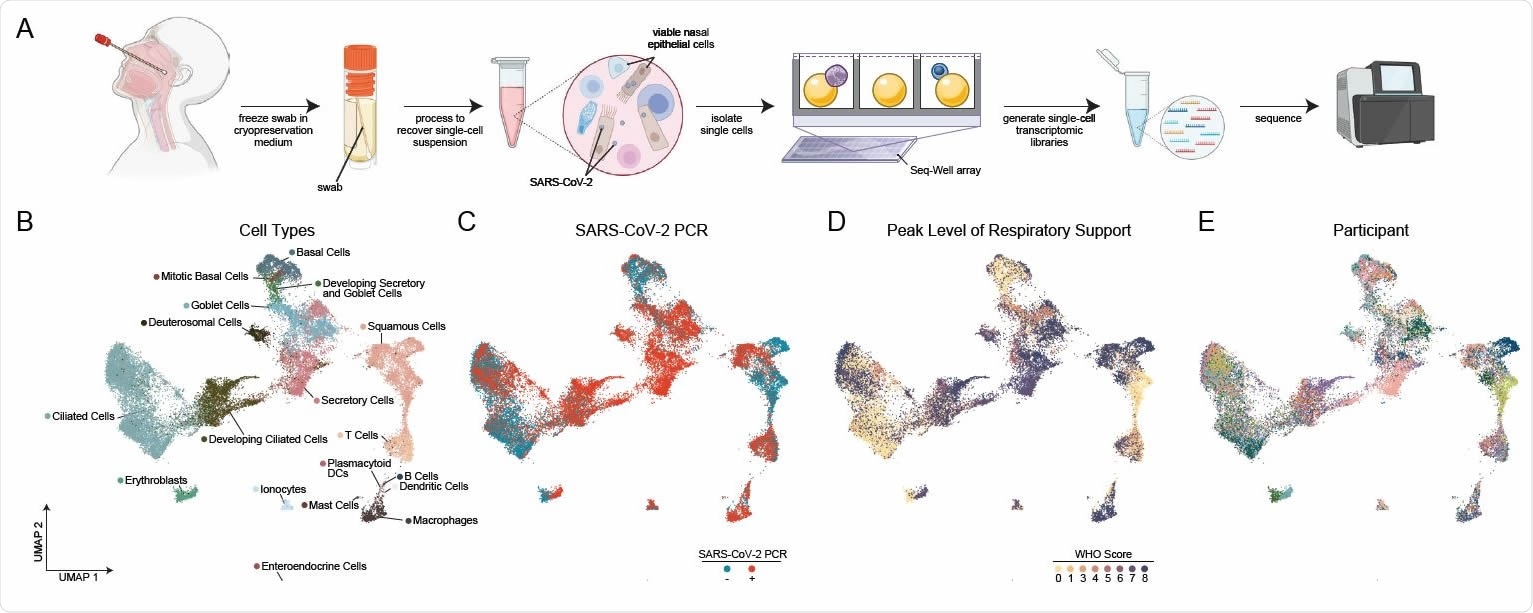
Cellular Composition of Human Nasopharyngeal Mucosa A. Schematic of method for viable cryopreservation of nasopharyngeal swabs, cellular isolation, and scRNA-seq using Seq-Well S3 (created with BioRender). B. UMAP of 32,588 single-cell transcriptomes from all participants, colored by cell type (following iterative Louvain clustering). C. UMAP as in B, colored by SARS-CoV-2 diagnostic PCR status. D. UMAP as in B, colored by peak level of respiratory support (WHO COVID-19 severity scale). E. UMAP as in B, colored by participant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
However, in many cases, infection is asymptomatic or confined to upper respiratory symptoms.
Ordovas-Montanes and colleagues say it is not yet clear which cell types in the human nasopharynx are the primary targets of SARS-CoV-2 infection.
“Few studies have directly addressed the impact of SARS-CoV-2 infection on the respiratory epithelium of the human upper airways, or examined how this may relate to aberrant inflammatory or antiviral signaling described in the periphery,” say the researchers.
“A clearer understanding of viral tropism, how the airway epithelium responds to infection, and the relationship to disease outcome may critically inform future therapeutic or prophylactic strategies,” they add.
What did the researchers do?
The team comprehensively analyzed cellular phenotypes within nasal mucosa during early SARS-CoV-2 infection by performing single-cell RNA-sequencing (scRNA-seq) of nasopharyngeal swabs collected from a large patient cohort.
The cohort included 35 COVID-19 patients with disease states ranging from ambulatory to critically ill and 23 healthy patients who did not have COVID-19.
The researchers simultaneously profiled both host and viral RNA and created a detailed map of epithelial and immune cell diversity.
What did they find?
Infection with SARS-CoV-2 led to significant changes in the upper respiratory epithelium.
A dramatic loss of mature ciliated cells was associated with secretory cell expansion and accumulation of deuterosomal cell intermediates and immature ciliate cells, potentially representing compensatory repopulation of lost ciliated epithelium.
“Deuterosomal cells and immature ciliated cells were considerably expanded among COVID-19 samples, suggesting interdependence between each of these compartments in maintaining epithelial homeostasis during viral challenge,” say the researchers.
Epithelial cells from individuals with mild or moderate COVID-19 showed broad induction of genes associated with antiviral and type I interferon responses.
By contrast, epithelial cells from individuals with severe lower respiratory symptoms showed dramatically blunted antiviral capacity. Severe COVID-19 was also characterized by mucosal recruitment of highly inflammatory myeloid cells – the primary sources of pro-inflammatory cytokines.
“This suggests an essential role for intrinsic, local epithelial immunity in curbing and constraining viral-induced pathology,” says Ordovas-Montanes and colleagues.
By characterizing cell-associated SARS-CoV-2 RNA, the researchers identified rare cells with RNA intermediates that were suggestive of active replication.
Heterogeneous responses in the epithelial compartment
The study also revealed significant diversity and heterogeneity among host cells containing SARS-CoV-2 RNA, including immature and interferon-responsive ciliated cells and specialized subsets of secretory, goblet, and squamous cells.
“Understanding whether heterogeneous responses in the epithelial compartment between individuals with COVID-19 underscores differences in disease manifestations will require larger cohort studies, with a focus on longitudinal responses following initial infection,” advises the team.
Finally, compared with uninfected cells, those containing SARS-CoV-2 RNA were enriched for genes known to be involved in susceptibility or response to infection.
What did the authors conclude?
The researchers say the study suggests that intrinsic defects in immune or epithelial antiviral responses within the nasal mucosa may predispose to severe disease via enhanced viral replication in the upper airway.
“Further, it suggests that there may be a clinical window in which severe disease can be subverted by focusing preventative or therapeutic interventions early within the nasopharynx, thereby bolstering antiviral responses and curbing pathological inflammatory signaling prior to the development of severe respiratory dysfunction or systemic disease,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ordovas-Montanes J, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.20.431155, https://www.biorxiv.org/content/10.1101/2021.02.20.431155v1
- Peer reviewed and published scientific report.
Ziegler, Carly G.K., Vincent N. Miao, Anna H. Owings, Andrew W. Navia, Ying Tang, Joshua D. Bromley, Peter Lotfy, et al. 2021. “Impaired Local Intrinsic Immunity to SARS-CoV-2 Infection in Severe COVID-19.” Cell 184 (18): 4713-4733.e22. https://doi.org/10.1016/j.cell.2021.07.023. https://www.cell.com/cell/fulltext/S0092-8674(21)00882-5?.