Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which caused the coronavirus disease 2019 (COVID-19) pandemic, gains entry to the host cell by binding to the angiotensin-converting enzyme 2 (ACE2). COVID-19 has repeatedly demonstrated its ability to infect an increasing number of mammalian species, such as tigers, mink, white-tailed deer, and many more.
Researchers based at the Rice University, Texas, USA, have now demonstrated a novel method of predicting a species' susceptibility to contracting COVID-19, based on the animal's structure of ACE2.
A pre-print version of the research paper is available to read in full on the bioRxiv*server.
What was the study
SARS-CoV-2 has always been presumed to have a zoonotic origin, either arising from bat or pangolin coronaviruses. While it is still unknown exactly which of these it first spread from, its ability for cross-species transmission is what has allowed the global COVID-19 pandemic to ensue, after being passed to humans at some point in late 2019.
Since the identification of the virus, multiple reported cases of COVID-19 have been confirmed in a number of domestic and wild mammal species. This has raised concerns that cross-species transmissibility may result in both local ecological outbreaks, but also that infected animals may re-transmit the virus back onto humans.
Previous research has suggested that pangolin coronaviruses were more likely to have been the ancestral virus for SARS-CoV-2, as pangolin ACE2 receptors in host cells to which the virus binds to are similar in both humans and pangolins. Additionally, experimental infection of white-tailed deer, which also share a similar ACE2 structure, have shown susceptibility to the disease.
Dr. Ryan Cheng and colleagues of the Rice University set out to therefore experimentally predict species that may be at risk of contracting COVID-19, based on the protein sequence of ACE2, to establish a theoretical list of species potentially at-risk to the virus.
The study
ACE2 topology is widely variable amongst mammal species, and frequently many species distantly related may share ACE2 receptors with very similar appearances.
Cheng and colleagues examined a selection of ACE2 topologies from 63 animals and assessed the potential of each of these to allow the SARS-CoV-2 virus to bind. To do this, they used the "energetic frustration" concept, a part of protein folding theory, as a proxy for binding affinity.
Essentially, the more dissimilar a protein receptor is to a subject trying to enter it, the more energy will be expelled as that subject attempts to bind. Therefore, a foreign protein or virus subject may be prevented from cell entry through a "kinetic trap," as it would take more energy to enter the cell than it can expend. More significant energetic frustration, therefore, represents lower ACE2 binding affinity, whilst minimal frustration represents higher affinity, allowing COVID-19 to enter a host cell.
Using human ACE2 as a template, an energy-landscape analysis was conducted on the models of the animal ACE2, measuring how favorable each receptor was to COVID-19 spike binding.
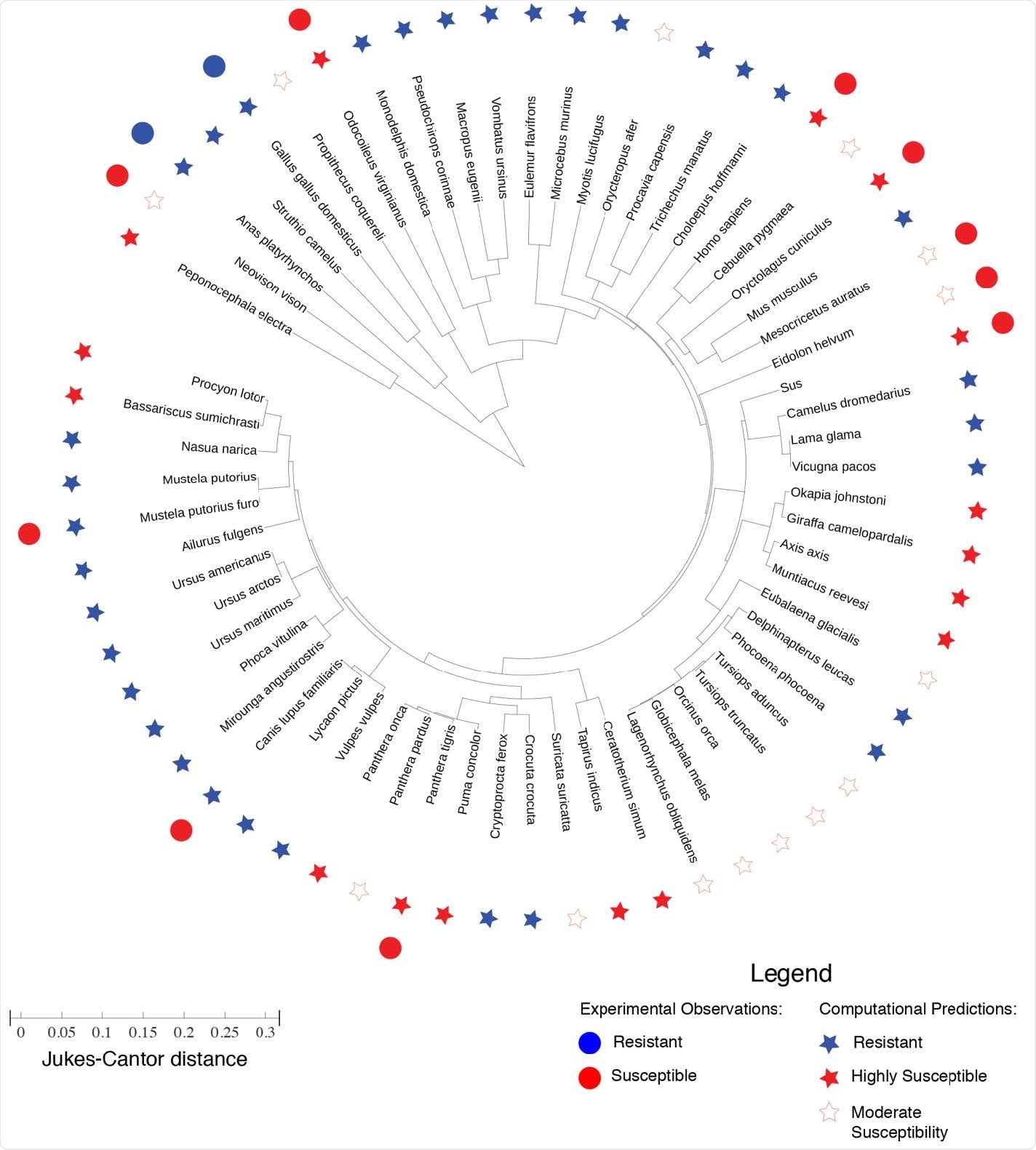
Phylogenic tree representing the evolutionary distance between ACE2 proteins of different species. The lengths in the radial direction denote the Jukes-Cantor distance as a measure of evolutionary distance between any two ACE2 proteins. The experimental observation of SARS-CoV-2 resistance/susceptibility are plotted alongside the computational predictions for resistance/susceptibility based on our frustration analysis of the a consistency between the computational predictions and the experimental observations for mouse (Mus musculus), chicken (Gallus gallus domesticus), duck (Anas platyrhynchos), mink (Neovison vison), bat (Eidolon helvum), Syrian golden hamster (Mesocricetus auratus), tiger (Panthera tigris), white-tailed deer (Odocoileus virginianus), European rabbit (Oryctolagus cuniculus), and pig (Sus scrofa). However, apparent inconsistencies are found for ferret (Mustela putorius furo) and dog (Canis lupis familiaris) —however, SARS-Cov-2 has only been observed to replicate in the upper respiratory tract of ferrets, and viral replication has been observed to be low in dogs.
What the study found
The researchers' findings suggest high susceptibility to SARS-CoV-2 infection for 16 of the 63 animals, with moderate susceptibility for a further 14. The remaining animals were predicted to have resistance to virus binding.
These findings corroborate well with experimental observations of SARS-CoV-2 infection in some of the animal species, however, with two exceptions: ferrets and dogs. The authors provide a plausible explanation for this:
"SARS-CoV-2 has only been observed to replicate in the upper respiratory tract of ferrets, and viral replication has been observed to be low in dogs."
Concluding remarks
The study's authors have demonstrated a potentially powerful tool for indicating species that may be at risk from SARS-CoV-2. Whilst right now the researchers only analyzed potential susceptibility through ACE2 – there are many more potential determinants at play most likely – it represents how an energy landscape-based analysis can be used to assess the interactions of ACE2 and SARS-CoV-2.
Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.