Researchers in the US have developed a novel multiplexed diagnostic platform based on CRISPR-based approaches and microfluidics - enabling robust and simultaneous surveillance of multiple viruses and viral variants. Currently, the paper is available on the medRxiv* preprint server pending peer review.
The ongoing coronavirus disease 2019 (COVID-19) pandemic quickly highlighted the need for high-throughput diagnostics in order to test large populations. However, early diagnostic efforts met several technical challenges that delayed our early response to the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Additionally, a number of challenges have emerged, including co-circulating human respiratory viruses that can cause symptoms quite similar to COVID-19, in addition to new SARS-CoV-2 variants with mutations that affect viral fitness, viral spread, and disease course.
Therefore, a perfect diagnostic method would come with surveillance capabilities to analyze hundreds of patient samples at the same time, unveil multiple viruses, recognize viral variants, and quantify viral load – but no such test currently exists in our diagnostic armamentarium.
From the technologies we actually do have, a reverse transcription-quantitative PCR can be considered high-throughput as it tests at least 88 samples, but for 1-3 analytes at a time. Furthermore, most novel CRISPR-based diagnostic approaches detect one or two targets per sample.
This is why a group of US researchers, led by Dr. Welch, Dr. Zhu, and Dr. Hua from the Broad Institute of MIT and Harvard, developed mCARMEN, which is a multiplexed, high-throughput and microfluidic diagnostic/surveillance platform for respiratory viruses and SARS-CoV-2 variants with the ability test 300-550 patient specimens in an eight-hour working day.
How mCARMEN was developed?
In order to develop mCARMEN into a clinically relevant technological solution, it was built on the shoulders of CARMEN version 1 by streamlining the workflow and incorporating commercially available Fluidigm microfluidics and instrumentation.
Then, mCARMEN was validated on 902 patient specimens to detect 9-21 human respiratory viruses or SARS-CoV-2 variant mutations with high agreement to comparator assays, which passed the performance criteria of the US Food and Drug Administration for all but one virus.
From the study, it was clear that mCARMEN workflow can process 188 patient samples for up to 23 respiratory viruses and variants in less than five hours, with costs that are lower than 10 USD per sample. Furthermore, in an academic setting, 21 human respiratory viruses were detected by mCARMEN, before the solution was taken in a hospital environment.
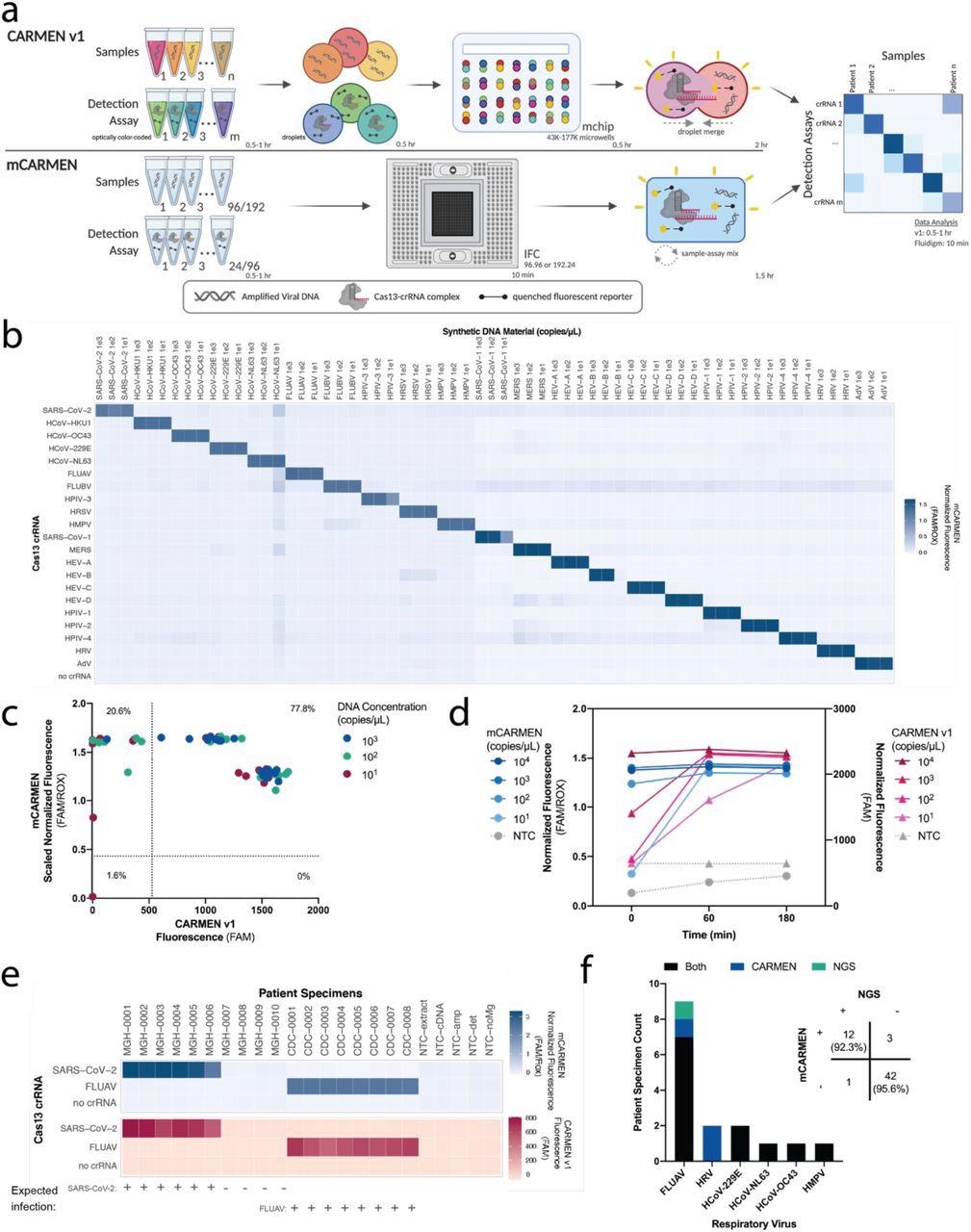 CARMEN implementation on Fluidigm achieves greater sensitivity quicker. a, Schematic of CARMEN v1 (top) and mCARMEN (bottom) workflows. b, Heatmap showing mCARMEN fluorescent data across 21 human respiratory viruses that were amplified using two separate primer pools. Synthetic DNA fragments were serially diluted from 103-101 copies/μL and added to Q5 amplification master mix. All samples were background-subtracted from NTC-noMg negative control. c, Concordance between CARMEN v1 and mCARMEN from b. Blue: targets at 103 copies/μL; green: targets at 102 copies/μL; red: targets at 101 copies/μL. d, Fluorescence kinetics of amplified SARS-CoV-2 DNA gene fragments from 104-101 copies/μL at 0, 60, and 180 minutes post-reaction initiation. Blue: mCARMEN; red: CARMEN v1. e, Testing 21 human respiratory virus panel on clinical specimens from 6 SARS-CoV-2 positive, 4 SARS-CoV-2 negative NP swabs, and 8 FLUAV positive specimens, collected prior to Dec. 2019, and 5 no target controls (NTCs). Heatmap shows fluorescent signals from SARS-CoV-2 crRNA, FLUAV crRNA and no crRNA control. Blue: mCARMEN; red: CARMEN v1. f, Concordance of mCARMEN and NGS on 58 suspected respiratory virus-infected patient specimens collected prior to Dec. 2019. Black: detected by both mCARMEN and NGS; blue: detected by mCARMEN only; green: detected by NGS only. mCARMEN values are shown as normalized fluorescence, FAM signal divided by passive reference dye, ROX, signal at 1 hour post-reaction initiation. CARMEN v1 values are shown as raw fluorescence, FAM signal at 3 hours. NTC, no target control; NTC-extract, no target control taken through extraction, cDNA synthesis, amplification, and detection; NTC-cDNA, no target control taken through cDNA synthesis, amplification, and detection; NTC-amp, no target control taken through amplification and detection; NTC-det, no target control taken through detection; NTC-noMg, no target control expected to have no fluorescent signal due to lack on Mg2+ needed to activate Cas13.
CARMEN implementation on Fluidigm achieves greater sensitivity quicker. a, Schematic of CARMEN v1 (top) and mCARMEN (bottom) workflows. b, Heatmap showing mCARMEN fluorescent data across 21 human respiratory viruses that were amplified using two separate primer pools. Synthetic DNA fragments were serially diluted from 103-101 copies/μL and added to Q5 amplification master mix. All samples were background-subtracted from NTC-noMg negative control. c, Concordance between CARMEN v1 and mCARMEN from b. Blue: targets at 103 copies/μL; green: targets at 102 copies/μL; red: targets at 101 copies/μL. d, Fluorescence kinetics of amplified SARS-CoV-2 DNA gene fragments from 104-101 copies/μL at 0, 60, and 180 minutes post-reaction initiation. Blue: mCARMEN; red: CARMEN v1. e, Testing 21 human respiratory virus panel on clinical specimens from 6 SARS-CoV-2 positive, 4 SARS-CoV-2 negative NP swabs, and 8 FLUAV positive specimens, collected prior to Dec. 2019, and 5 no target controls (NTCs). Heatmap shows fluorescent signals from SARS-CoV-2 crRNA, FLUAV crRNA and no crRNA control. Blue: mCARMEN; red: CARMEN v1. f, Concordance of mCARMEN and NGS on 58 suspected respiratory virus-infected patient specimens collected prior to Dec. 2019. Black: detected by both mCARMEN and NGS; blue: detected by mCARMEN only; green: detected by NGS only. mCARMEN values are shown as normalized fluorescence, FAM signal divided by passive reference dye, ROX, signal at 1 hour post-reaction initiation. CARMEN v1 values are shown as raw fluorescence, FAM signal at 3 hours. NTC, no target control; NTC-extract, no target control taken through extraction, cDNA synthesis, amplification, and detection; NTC-cDNA, no target control taken through cDNA synthesis, amplification, and detection; NTC-amp, no target control taken through amplification and detection; NTC-det, no target control taken through detection; NTC-noMg, no target control expected to have no fluorescent signal due to lack on Mg2+ needed to activate Cas13.
Maximizing the diagnostic relevance
In order to enhance the clinical diagnostic significance of mCARMEN and meld it with surveillance technology requirements, the researchers further augmented its multiplexing capabilities by discerning between mutations in viral variants found in patient specimens, as well as by quantifying viral genomic copies.
Moreover, they have developed a SARS-CoV-2 variant identification panel, which they have immediately used to detect the Omicron (B.1.1.529) variant during a case surge in the Boston area without any changes to the assay.
The assay was also adapted for dual Cas12 and Cas13 enzyme detection by taking into account differing protein kinetics, which is a current trend in CRISPR diagnostics. The researchers have also expanded the quantifiable concentration range to 5-6 orders of magnitude, which is similar to reverse transcription-quantitative PCR.
The aforementioned mCARMEN applications may provide a more holistic diagnosis to the patient. However, there are particular challenges before anticipating a large-scale roll-out (such as specimen degradation issues and funding support).
Implications for infectious disease surveillance
Even though further work will be needed to completely establish mCARMEN for clinical usage, this technological solution can function as an optimal single technology platform that comes with diagnostic and surveillance capabilities for detecting respiratory pathogens and viral variants.
“There is currently no other diagnostic technology that combines multiplexed pathogen testing with variant tracking and is highly scalable and amenable to clinical laboratory settings”, say study authors in this medRxiv paper. “We have taken significant steps to streamline assay workflow while enhancing sensitivity and not sacrificing specificity”, they add.
In a nutshell, the study has demonstrated how this approach can offer a very comprehensive and high-throughput diagnostic modality while at the same time maintaining a high level of clinical relevance unmatched by other types of antigen-based or nucleic acid-based diagnostics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Welch, N.L., Zhu, M., Hua. C., et al. (2021). Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and SARS-CoV-2 variants, medRxiv, https://doi.org/10.1101/2021.12.14.21267689, https://www.medrxiv.org/content/10.1101/2021.12.14.21267689v1
- Peer reviewed and published scientific report.
Welch, Nicole L., Meilin Zhu, Catherine Hua, Juliane Weller, Marzieh Ezzaty Mirhashemi, Tien G. Nguyen, Sreekar Mantena, et al. 2022. “Multiplexed CRISPR-Based Microfluidic Platform for Clinical Testing of Respiratory Viruses and Identification of SARS-CoV-2 Variants.” Nature Medicine 28 (5): 1083–94. https://doi.org/10.1038/s41591-022-01734-1. https://www.nature.com/articles/s41591-022-01734-1.