The increase in high throughput sequencing technologies has broadened the known diversity of the virosphere. These technologies have enhanced the frequency and accuracy of viral surveillance. In an Amazonian metropolitan region, scientists have been studying the viral communities associated with the human-animal interface. Now, a recent study in the journal Viruses focused on the identification of a novel rodent-borne arterivirus.
Viruses belonging to the Arteriviridae family have been characterized as spherical, enveloped, single-stranded, and positive-sense multi-cistron RNA genomes. In the context of their genomic organization, a marginal variation in the open reading frame (ORF) was observed among all species belonging to the Arteriviridae family. Conserved domains have been identified that include ORFs 1a (3C-like protease-3CLpro) and 1b (RNA-dependent RNA polymerase-RdRp). These are standard markers for genetic distance indexes and phylogenetic inferences used for taxonomic differentiation within the family.
Non-human mammalian viruses within wide-ranging hosts, such as rodents, shrews, horses, hedgehogs, and possums, which have yet not been assigned to any species, are known as arterivirids. The majority of arteriviruses, such as simian hemorrhagic fever virus (SHFV) and equine arteritis virus (EAV), are pathogens of veterinary importance. However, lactate dehydrogenase elevating virus (LDV), which belongs to the Arteriviridae family, infect mice.
Most recently described rodent arteriviruses have African, Asian, or European origins. These viruses were identified from samples collected from Ukraine, China, Cameroon, Mozambique, and Tanzania through metagenomic studies.
About the Study
The current study is part of a large surveillance study to explore the viral communities associated with wild mammals at the human–animal interface. Tissue samples were collected from wild mammals in Santa Bárbara do Pará, Brazil. For this study, animals were captured using Tomahawk and Sherman traps in forest areas with close human habitations and agricultural practices. A total of 39 mammals, 12 rodents, 18 marsupials, and 9 chiropterans were captured.
Blood and serum samples were collected from the captured animals. These animals were then anesthetized and euthanized. Subsequently, tissue samples from lymph nodes, spleen, heart, and lungs, were collected. Tissue samples of rodents were pooled and analyzed. Oecomys sp. was identified from rodents based on morphological characteristics. The rodents did not present any apparent signs of illness.
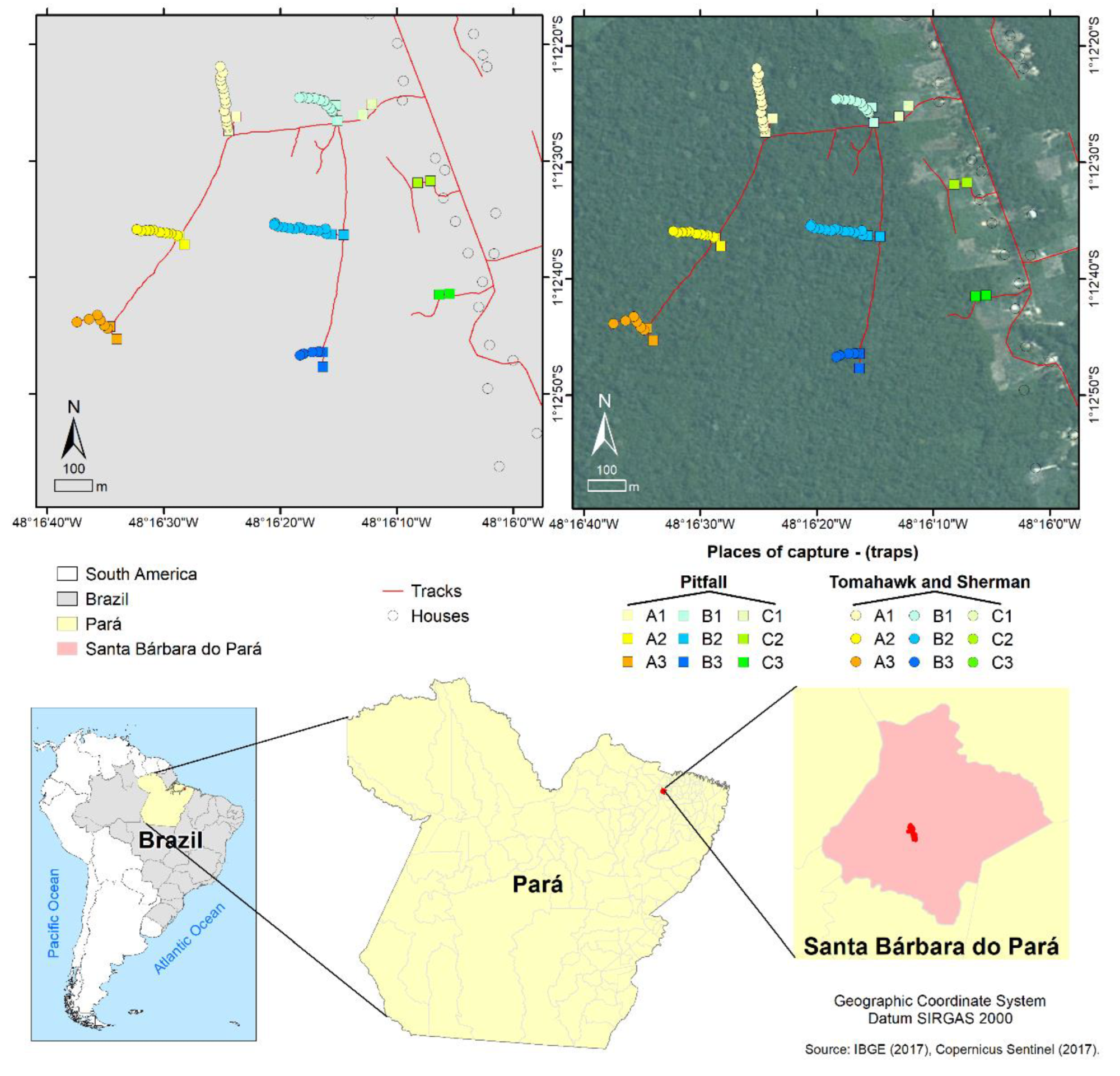 Map of the collection area in the Expedito Ribeiro village in Santa Bárbara do Pará municipality (Pará state, Brazil) and its location in the Brazilian territory. Traps were placed in the open field surrounding the human habitations (A1–A3), the forest fragment border (B1–B3), and its most interior region (C1–C3). The IBGE (2017) and Copernicus Sentinel (2017) databases were consulted to prepare the maps.
Map of the collection area in the Expedito Ribeiro village in Santa Bárbara do Pará municipality (Pará state, Brazil) and its location in the Brazilian territory. Traps were placed in the open field surrounding the human habitations (A1–A3), the forest fragment border (B1–B3), and its most interior region (C1–C3). The IBGE (2017) and Copernicus Sentinel (2017) databases were consulted to prepare the maps.
Study Findings
An almost complete genome was retrieved, which contained four final contigs related to known arterivirids. The genomic sequence revealed that the virus was a member of the Arteriviridae family.
ORF1a and ORF1b, which are the two most prominent ORFs of arterivirid genomes, were identified. ORFs expressing the non-structural proteins (NSPs) and structural proteins (envelope (E), glycoproteins (GP2 to GP5), membrane (M), and nucleocapsid (N) were detected. InterProScan search was used to detect three cysteine-protease domains of NSP1 and NSP2, characteristic of arteriviruses, in the predicted sequence of polyprotein 1a.
The complete mitochondrial genome of the host was recovered, which showed a 99.38% similarity with Oecomys paricola. This virus was tentatively named after the host genus, Oecomys arterivirus 1 (OAV-1). In the phylogenetic analysis, this novel arterivirus was placed with a high support bootstrap value at the most basal branch in the clade of porcine and rodent arterivirids. This corresponded to the Variarterivirinae subfamily clade, the most proximal taxon.
Even though the mean distance between OAV-1 and members of the Variarterivirinae subfamily slightly exceeds the maximum intragroup distance for the subfamily, this taxon is most related to the virus. The distance values between OAV-1 and the Variarterivirins are entirely out of the intergroup range.
This study revealed that OAV-1 is highly divergent on both amino acid and nucleotide levels, which indicated that it could be a part of a distinct clade of arteriviruses. This clade could be exclusive to the region. Based on distance analysis, this virus is a divergent representation of a new genus in the Variarterivirinae subfamily.
Conclusions
The discovery of novel arteriviruses may challenge the current taxonomic classification. Although arteriviruses are non-human viruses, their significance as wildlife and livestock pathogens must not be neglected. The Brazilian arboreal rice rat, Oecomys paricola, was identified as the host of OAV-1. Since the increased contact between domestic and wild species favors viral spillover, further surveillance studies are required to uncover the diversity and coevolutionary patterns of the Arteriviridae family in the Amazon.
usechatgpt init success