The Human Intestinal Bacteria Collection (HiBC) offers scientists access to over 300 live gut bacterial strains, many never studied before, empowering research into inflammation, immunity, and future microbiome-based therapies.
 HiBC: a publicly available collection of bacterial strains isolated from the human gut. Image Credit: Tatiana Shepeleva / Shutterstock
HiBC: a publicly available collection of bacterial strains isolated from the human gut. Image Credit: Tatiana Shepeleva / Shutterstock
Very few strains out of the multitude of bacteria that inhabit the human gut are accessible to scientific research, and a vast majority remain largely unknown. To address this dearth of knowledge, researchers from Germany unveiled a new and publicly available collection of human gut bacteria, called the Human Intestinal Bacteria Collection (HiBC), in a recent study published in the journal Nature Communications. Notably, the HiBC project also undertook the formal taxonomic description and naming of new species and systematically deposited every strain in international public culture collections—an uncommon practice that ensures global researcher access. This collection also contains 29 never-before-seen species and offers powerful tools to understand how gut microbes affect human health and disease.
The gut microbiome
The human gut is home to trillions of bacteria that influence digestion, immunity, and even mental health. While advances in deoxyribonucleic acid (DNA) sequencing have identified many of these microbes, most remain poorly understood because they have never been grown in a lab. This limits the ability to study their function or test how they affect the body.
Past efforts to isolate gut bacteria have led to valuable collections, but few strains have been made publicly accessible. Moreover, the accessible ones often lack key information like genome quality, growth conditions, and host origin. Many potentially important bacterial species are known only from their genetic signatures and not as living organisms that can be studied directly.
These challenges create gaps in microbiome research, especially in understanding how bacteria interact with human cells or respond to therapies. To unlock these insights, scientists need well-characterized and readily available bacterial strains. This would allow direct experimentation to reveal how specific microbes influence health or disease.
The current study
The researchers first collected human stool samples to build a comprehensive collection of gut bacteria after obtaining ethical approval and participant consent. Samples were stored anaerobically to preserve oxygen-sensitive microbes. Within 24 hours, they were processed in a controlled oxygen-free environment to maintain the integrity of the bacterial communities.
The researchers used two main isolation techniques: the classical method, which involves dilution of fecal samples and spreading them on agar plates to grow bacterial colonies, and a second method, which uses a single-cell dispensing system to isolate individual bacteria more precisely.
Each isolated strain was initially identified using mass spectrometry. If identification was unclear or a novel species was suspected, DNA was extracted for genome sequencing. Genome sequencing used Illumina platforms, followed by quality filtering, adapter removal, and assembly. Only high-quality genomes with over 90% completion and less than 5% contamination were included.
The study also measured the ecological relevance of strains by comparing their 16S ribosomal ribonucleic acid (rRNA) genes to global microbiome datasets. This helped confirm how widespread and potentially important each strain is. Moreover, all information, including genome data, plasmid sequences, and growth conditions, was made publicly accessible through the HiBC website.
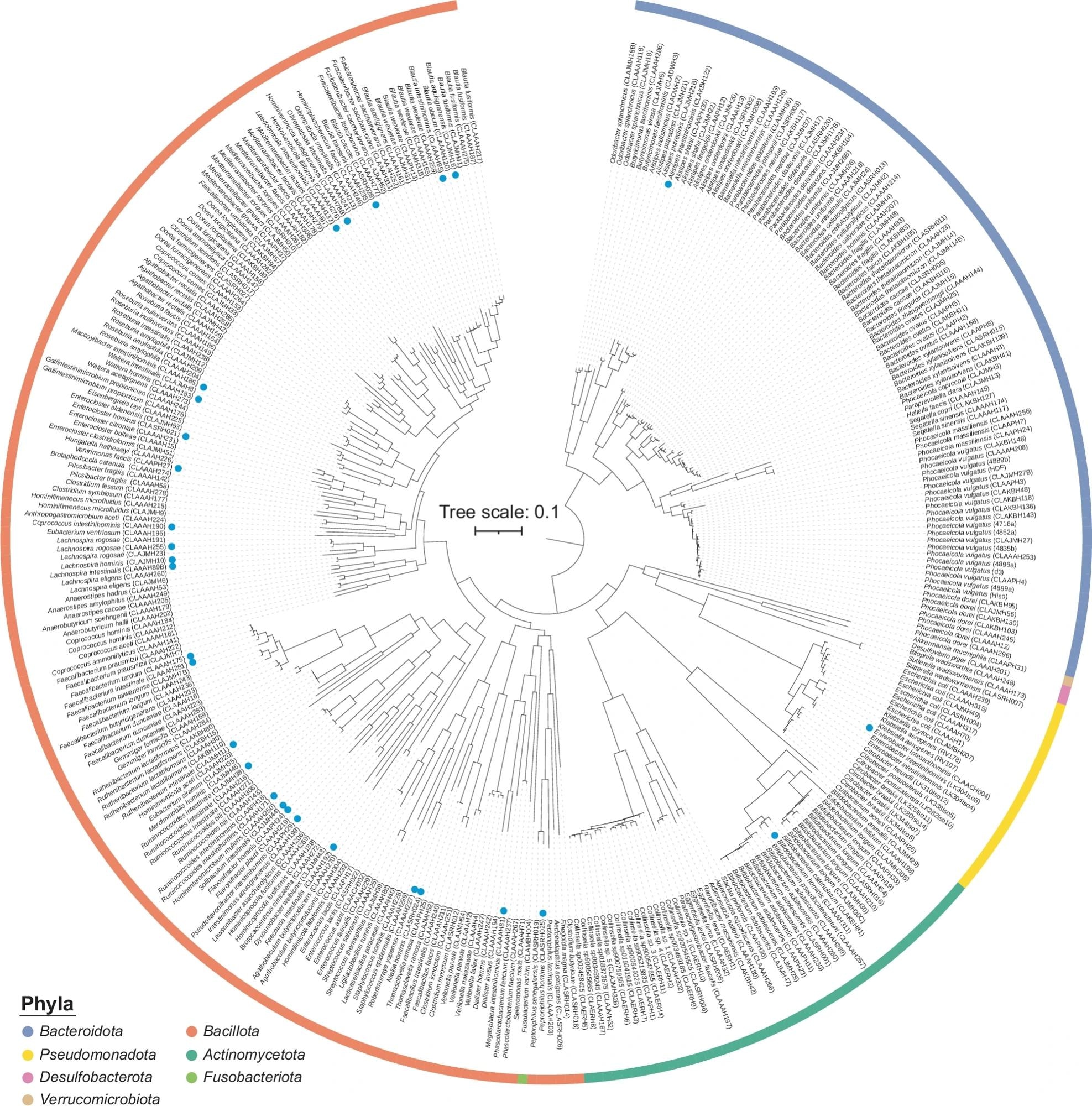 Tree based on all 340 genomes, generated using Phylophlan. Phyla are indicated with colours. The Bacillota are split due to the placement of Fusobacteriota, which separated strains assigned to ‘Bacillota_A’ by GTDB, however, this is dependent on the method used for tree creation. The potential need for splitting the phylum Bacillota is therefore independently supported by the Phylophlan tree and GTDB. Blue circles identify strains belonging to the 29 novel species that are taxonomically described in this paper.
Tree based on all 340 genomes, generated using Phylophlan. Phyla are indicated with colours. The Bacillota are split due to the placement of Fusobacteriota, which separated strains assigned to ‘Bacillota_A’ by GTDB, however, this is dependent on the method used for tree creation. The potential need for splitting the phylum Bacillota is therefore independently supported by the Phylophlan tree and GTDB. Blue circles identify strains belonging to the 29 novel species that are taxonomically described in this paper.
Key findings
The study reported that the HiBC comprises 340 bacterial strains representing 198 distinct species, including 29 species never previously described. This makes the HiBC one of the most comprehensive and accessible gut bacteria collections to date. The team’s systematic effort to deposit every strain into public culture collections, such as DSMZ, BCCM, and JCM, also helped overcome a major limitation in microbiome research, ensuring that both living isolates and their metadata are available for future studies.
The isolates in the collection belonged to seven major bacterial phyla commonly found in the human gut, with especially high representation from the Bacillota and Bacteroidota groups. The newly described species included novel types of Faecalibacterium, known for their anti-inflammatory effects, and other species associated with both health and gastrointestinal disease.
Nearly half of the strains contained plasmids, which are mobile genetic elements that can influence bacterial traits such as antibiotic resistance and colonization ability. Among the 266 reconstructed plasmids, some were identified as "megaplasmids" that were over 100,000 base pairs long. One such plasmid, pMMCAT, was found to boost surface adhesion, a trait that may help bacteria colonize the gut.
Additionally, the analysis revealed that several of the novel species are commonly found across human populations. Some strains were detected in over 90% of individuals sampled in global studies, while others were present at lower but still significant prevalence. This suggests that these microbes may play important roles in gut health.
The study further linked specific bacterial features with diseases such as Crohn’s disease and ulcerative colitis. Furthermore, certain proteins encoded by the isolated genomes were found to be associated with disease status, with some more prevalent in healthy individuals and others in patients. This highlights the complex relationship between gut microbes and disease, suggesting that these microbes may contribute to disease development or protection.
Conclusions
Overall, the HiBC offers a vital new resource for studying gut microbes, providing both living strains and comprehensive genomic data. It enables more precise functional and mechanistic studies and significantly advances microbiome research by supporting the experimental validation of microbe–host interactions.
Additionally, the HiBC makes data on gut bacteria and their genetic information accessible, opening doors to better diagnostics, therapeutics, and improving our general understanding of the human microbiome.
Journal reference:
- Hitch, T.C.A., Masson, J.M., Pauvert, C. et al. (2025). HiBC: a publicly available collection of bacterial strains isolated from the human gut. Nature Communications 16, 4203. DOI: 10.1038/s41467-025-59229-9, https://www.nature.com/articles/s41467-025-59229-9