In drug discovery and research projects, an important focus is the determination and characterization of binding interactions between proteins and low molecular weight (LMW) ligand interactions. Isothermal titration calorimetry (ITC) is a technique used to directly determine the amount of heat released or absorbed during a binding event. This method allows for the study of interactions between proteins and small molecules in solution, without immobilization or labeling being required. The ITC technique is often used to characterize the variations in enthalpic and entropic contributions to binding of new ligands.
Wide affinity range is one of the challenges associated with the quantification and characterization of binding interactions. In a standard drug discovery project, binding affinity values (KD) of small compounds binding to a target protein can range from low millimolar and subnanomolar. An equally wide affinity range is applicable for academic research projects, depending on the biochemical system being analyzed.
Using properly designed experiments, along with high-sensitivity ITC instruments can simply the characterization of binding interactions significantly. The MicroCal PEAQ-ITC, a new ITC system, has been designed to improve signal stability, mixing and signal-to-noise ratio (SNR) characteristics, as shown in Figure 1.
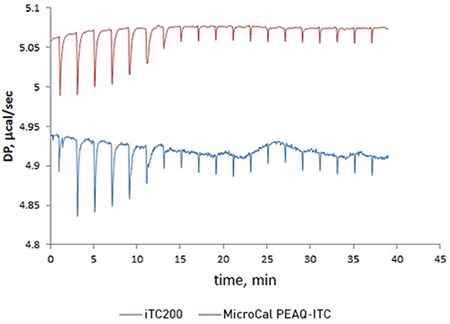
Figure 1. Overlay of raw ITC data for the titration of 20 μM acetazolamide (ACZA) into 2 μM bovine carbonic anhydrase II (bCAII) using the MicroCal PEAQ-ITC (top) and MicroCal iTC200 (bottom) systems.
These changes, together with the sophisticated experimental design integrated in the user friendly data analysis software, enable experiments to be optimized for the analysis of challenging binding interactions. This article describes the advantageous features of the MicroCal PEAQ-ITC instrument and software for studying binding interactions via competitive and direct titrations.
MicroCal PEAQ-ITC Instrument with Improved Data Quality
To compare the new MicroCal PEAQ-ITC system with its predecessor, the MicroCal iTC200, a titration of acetazolamide with bovine carbonic anhydrase II was carried out. The data obtained from two sets of experiments demonstrated that the MicroCal PEAQ-ITC has a short term noise that is around five times lower. This improved data quality boosts confidence when studying challenging data with low heats when the sample is precious or when low concentrations are required to accurately measure low-nanomolar affinity interactions.
Broad Range of Binding Affinities
In this experiment, the applicability of the new PEAQ MicroCal ITC system for characterizing binding interactions is demonstrated by carrying out titrations of human carbonic anhydrase I (hCAI) and bovine carbonic anhydrase II (bCAII) with several inhibitors. These examples span a wide range of enthalpies (approximately6 to -15 kcals/mol) and KDs (0. 320 mM to about 1 nM). Figure 2 shows the typical data sets, and Table 1 summarizes the results.
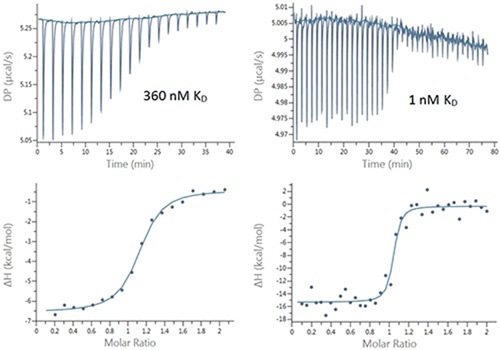
Figure 2. Raw (top) and integrated heat (bottom) plots for the titration of 200 μM Furosemide into 20 μM bCAII (left) using 2 μL injections and 25 μM ethoxzolamide into 2.5 μM bCAII (right) using 1 μL injections.
|
Protein
|
cmpd, n=3
|
N (sites)
|
Error (N), %
|
KD (M)
|
pKD
|
Error (KD), %
|
ΔH (kcal/ mole)
|
Error (ΔH), %
|
|
hCAI
|
Sulfanilamide
|
1.0
|
Fixed
|
3.2E-04
|
3.5
|
16
|
-5.9
|
15
|
|
Ethoxzolamide
|
1.0
|
1.0
|
1.9E-08
|
7.7
|
7
|
-8.7
|
1
|
|
bCAII
|
Furosemide
|
1.0
|
1.7
|
3.6E-07
|
6.4
|
18
|
-6.3
|
3
|
|
Acetazolamide
|
0.9
|
0.6
|
1.8E-08
|
7.7
|
3
|
-12.6
|
3
|
|
Ethoxzolamide
|
0.9
|
0.3
|
4.4E-10
|
9.4
|
95
|
-14.4
|
3
|
|
Ethoxzolamide Vinj=1 uL
|
1.0
|
1.0
|
1.3E-09
|
8.8
|
76
|
-15.1
|
1
|
Table 1. Summary of the results for the titrations (n=3) of hCAI and bCAII with a series of inhibitors.
First, ligands were injected into the target proteins at the following concentrations:
- 2.5 mM sulfanilamide into 35 μM hCAI
- 25 μM ethoxzolamide into 2.5 μM bCAII
- 50 μM ethoxzolamide into 5 μM hCAI
- 50 μM acetazolamide into 5 μM bCAII
- 200 μM furosemide into 20 μM bCAII
Next, experiments were carried out in PBS buffer, 2% v/v at 25°C. All interactions were analyzed using a standard protocol of injecting 18, 2 μL aliquots of ligand into the cell’s protein solution. For interactions with affinities of 18 nM and weaker, errors in KD were all below 20%, with some less than 10%. Errors in the stoichiometry and enthalpy data were also less than 3%, with the exception of sulfanilamide: hCAI interaction, where the N was fixed and enthalpies changed by 15%. Such measurements demonstrate the high reproducibility of the data that can be achieved when using the MicroCal PEAQ-ITC system, even when the protein concentrations were as low as 2.5 μM.
As the tightest binder, ethoxzolamide displayed the highest variation of KD value in triplicate experiments, as shown in Table 1, due to the relatively high C-value and a transition range that was less well defined and represented by just two experimental data points on the unique binding isotherm.
However, the 94% error is often adequate for these tight interactions and a certain level of confidence can be placed in the data. The low SNR and high precision pipette in the MicroCal PEAQ-ITC system meant it was possible to use smaller, 1μl injection volumes. Using this technique, the transition region can be populated (Figure 2) with more data points to improve the quality of the affinity data. This was indicated by the small but quantifiable decrease in the error in the KD value to 76%, which further increases confidence in the affinity data. Figure 3 illustrates these data in terms of pKD, which may be more appropriate for comparing affinity data.
* C-Value is defined as C = N*[Cell]/KD, where [Cell] is molar concentration of macromolecule in the cell, KD is the equilibrium dissociation constant, and N denotes the number of binding sites on the macromolecule.
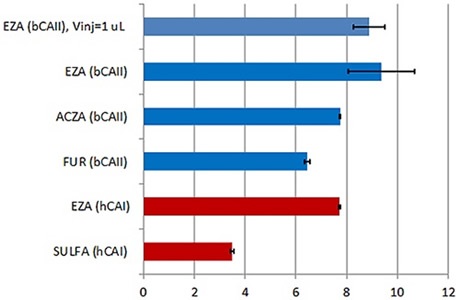
Figure 3. Summary of the KD values obtained with the new MicroCal PEAQ-ITC instrument and analysis software for a series of LMW inhibitors of hCAI (red) and bCAII (blue). All the data were collected with 18 × 2 μl injections of ligand into protein. In addition, KD value obtained in titration with 38 × 1 μl injections (first injection of 0.4 μl) is also reported for bCAII interaction with ethoxzolamide (EZA). To facilitate the comparison, the KD values on the figure are expressed as pKD. Error bars indicate errors in units of pKD (errors in % of KD are given in Table 1). Abbreviations: EZA –ethoxzolamide, FUR-furosemide, ACZA-acetazolamide, SULFA-sulfanilamide, bCAII –bovine carbonic anhydrase II and hCAI – human carbonic anhydrase I.
Competition Titration as a Technique for Quantitating Low Nanomolar Binding
Based on the data described above, it is clear that as the affinities become tighter, errors in the determination of KD increase. In such situations, it would be appropriate to use a competition titration. During a competition experiment, the target protein is injected with the strong binding ligand in the presence of a competitive, weaker compound. When the weaker inhibitor is not present, the ITC titration lacks the adequate number of data points in the transition region needed for fitting to a unique binding isotherm. An example of this is illustrated in Figure 4A, where 200 μM of ethoxzolamide was injected into 20 μM bCAII in 2 μL aliquots.
By using the competition experiment design tool the MicroCal PEAQ-ITC provides, (Figure 5), it is possible to identify the experimental conditions that would ensure an adequate number of data points in the transition range and at a sufficiently high heat value for good SNR. A set up with 100 μM furosemide added to the protein solution was chosen and the titration that resulted is illustrated in Figure 4B.
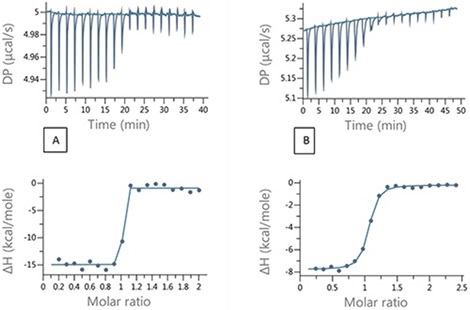
Figure 4. Raw and integrated ITC data for (A) direct titration of 200 μM EZA into 20 μM bCAII and (B) Competition titration 200 μM EZA into 20 μM bCAII and 100 μM FUR.
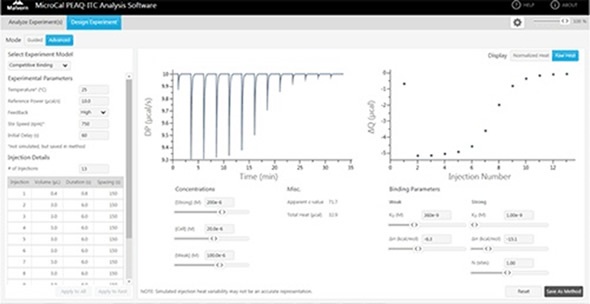
Figure 5. The competition experiment design tool in the MicroCal PEAQ-ITC.
The binding parameters for the tight binder obtained from the competition titration agreed with the data obtained from direct titration, as shown in Table 2. This study shows the effectiveness of using the design tool and the competition experiments for determining tight binding interactions.
|
Experimental setup
|
KD (nM)
|
error(KD), %
|
ΔH (kcal/mol)
|
error(DH), %
|
|
Direct titration
|
0.4
|
95
|
-14.4
|
3
|
|
Competition titration
|
0.4
|
30
|
-13.9
|
6
|
Table 2. Binding parameters of bCA II interaction with EZA obtained in direct titration of 25 μM EZA into 2.5 μM bCA II and in a competition titration of 200μM EZA into 20μM bCA II premixed with 100 μM FUR, Vinj=2 μL.
Conclusion
The data demonstrated here show the quality of data that can be obtained when using the Microcal PEAQ-ITC system. The system has a high SNR and can be used to determine a wide affinity range. The interaction between bCAII and EZA, which was the tightest example measured, has an affinity of 1nM with only two-fold variability. This level of repeatability is exceptional for any method used to quantify nanomolar affinities. Here, the micromolar interactions measured had errors of just 20% or better, representing exceptional repeatability for KD determination.
Competition experiments can be used to extend the affinity range that the ITC technique can be used for into the high picomolar range. However, one disadvantage of this approach is the complexity of the experimental design. This MicroCal PEAQ-ITC analysis software has a simulation tool that can simplify the design and analysis. Besides improving SNR, the MicroCal PEAQ-ITC system features fully automated data analysis, thereby reducing analysis time and user subjectivity in the assessment of data quality. For each experiment, data quality can be determined and fitting is performed in just a few seconds, allowing for analysis of large data sets of 50 or more experiments in a matter of seconds.
About Malvern Panalytical

Malvern Panalytical provides the materials and biophysical characterization technology and expertise that enable scientists and engineers to understand and control the properties of dispersed systems.
These systems range from proteins and polymers in solution, particle and nanoparticle suspensions and emulsions, through to sprays and aerosols, industrial bulk powders and high concentration slurries.
Used at all stages of research, development and manufacturing, Malvern Panalytical’s materials characterization instruments provide critical information that helps accelerate research and product development, enhance and maintain product quality and optimize process efficiency.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.