Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a betacoronavirus related to human SARS-CoV and bat SARS-like CoVs. The genome of SARS-CoV-2 reveals similarities to SARS-CoVs and key differences that may explain the increased transmissibility of SARS-CoV-2 compared to SARS-CoV.
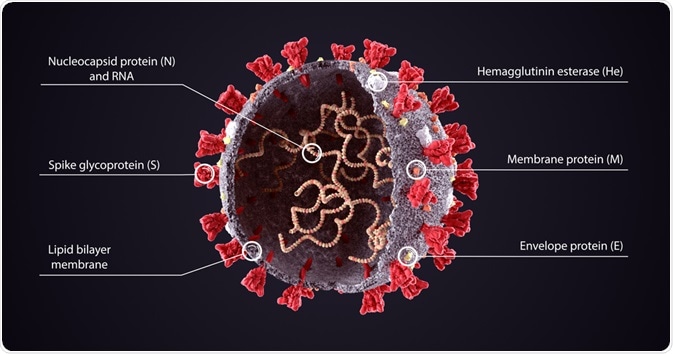 Image Credit: Orpheus FX/Shutterstock.com
Image Credit: Orpheus FX/Shutterstock.com
SARS-CoV-2
Severe acute respiratory syndrome coronavirus (SARS-CoV-2), formerly known as 2019-nCoV, is the virus that causes COVID-19. It is a lineage b beta-coronavirus belonging to the coronaviridae family of the order nidovirales (see Phylogenetic Tree of SARS-CoV-2). SARS-CoV-2 is a Baltimore class IV positive-sense ssRNA virus and is thought to have zoonotic origins.
SARS-CoV and other coronaviruses
Of the different types of coronaviruses, alpha- and beta-coronaviruses tend to infect mammals, whereas gamma- and delta-coronaviruses tend to infect birds. Of all those that infect humans, there are currently 7 known human CoVs: HCoV-NL63, HCoV-229E (both alphacoronaviruses), and HCoV-OC43, HCoV-HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 (beta coronaviruses).
SARS-CoV-2 shares the highest overall genome sequence similarity to the bat coronavirus RaTG13. However, the RBD shares more sequence similarity with the pangolin coronavirus SARS-CoV(MP489/Guandong/2019), which binds to ACE2. This suggests evidence of a recombination event in the evolution of SARS-CoV-2 due to animal mixing before evolving to infect humans.
All coronaviruses contain 26,000-32,000bp genomes with a variable 6-11 open reading frames (ORFs), which encode non-structural proteins as well as structural proteins: spike glycoprotein (S), a small envelope protein (E), matrix protein (M) & nucleocapsid protein (N).
SARS-CoV-2 is related to SARS-CoV, the virus that causes SARS, in addition to SARS-like bat CoVs (RaTG13/CoVZC45/CoVZCX21). All bind to the ACE2 receptor due to their similar S domain genomic sequences. Furthermore, a large proportion of SARS-CoV and SARS-CoV-2 genomes are identical. However, notable differences occur in key locations such as the absence of 8a protein and longer 8b protein in SARS-CoV-2.
In one analysis, there were 380 amino acid substitutions between SARS-CoV-2 and SARS/SARS-like viruses, specifically located in nsp2 & nsp3. 6 substitutions were found in the RBD at amino acid location 357-528, with a further 4 substitutions in the C-terminus region of the S1 domain.
However, no substitutions were found in nsp7, nps13, E, M, and accessory proteins p6 and 8b. This suggests that the differing RBD domain substitutions are probably behind the increased pathogenicity and transmissibility between SARS-CoV-2 and SARS-CoV.
SARS-CoV-2 possesses an intact open reading frame 8 without the 29-nt deletion found in a majority of human SARS-CoV. Furthermore, SARS-CoV-2 contains a distinct proteolytically sensitive activation loop (polybasic furin cleavage site) at the S1-S2 junction that is not typically found in other human lineage b beta coronaviruses.
However, this is a feature of several other animal and human coronaviruses, such as HKU1 (lineage a). Compared to SARS-CoV, the addition of such a cleavage site is thought to enhance cell-cell fusion. Many studies have shown that the polybasic furin cleavage site in SARS-CoV-2 promotes cell-cell fusion and infectivity.
MERS-CoV diverges from SARS-CoV & SARS-CoV-2 at the RBD as MERS-CoV binds to DDP4. Furthermore, encoded proteins pp1ab, pp1a, E, M, accessory protein 7a, and N genes vary considerably between SARS-CoV/CoV-2 genomes.
SARS-CoV-2 and influenza viruses
Other major respiratory viruses include Influenza viruses, which belong to the Orthomyxoviridae family of the orthornavirae kingdom. Influenza A and B types infect humans, mammals & birds and cause annual flu epidemics, whereas type C normally causes only mild illness in humans, and D is not known to cause disease in humans, primarily affecting cattle. All major flu pandemics have been caused by Type A influenza viruses.
Influenza viruses are negative-sense RNA viruses compared to positive-sense coronaviruses. Seasonal flu vaccines are designed to incorporate recent seasonal Influenza type A strains and offer little to zero protection against coronaviruses.
Aside from the vast genomic differences between lineage b beta coronaviruses and type A Influenza viruses, the presence of an efficient cleavage site in SARS-CoV-2, and haemagglutinin (HA) on Influenza strains, may be the causative factor being rapid viral replication and transmission and increased pathogenicity of both viruses.
As SARS-CoV-2 has a furin recognition motif insertion (PRRA) at the S1-S2 intersection not present in the currently recognized closest relatives SARS-CoV-2, it may be that SARS-CoV-2 acquired this from the first human-human transmission or through a yet unidentified bat or pangolin SARS-like CoV. Recent research has shown that furin sites have naturally occurred many times during coronavirus evolution.
Summary
In summary, SARS-CoV-2 shares a high degree of genomic similarity between other beta coronaviruses, including human SARS-CoV and bat SARS-like-CoVs/RaTG13. Whilst their genomes are largely similar, key genomic differences are suggesting a different evolution of SARS-CoV-2, most likely involving batSARS-like-CoVs.
Understanding the genomic differences will help scientists develop highly effective vaccines against SARS-CoV-2 and learn how viruses evolve.
COVID-19 Symposium: SARS-CoV-2 Genome Sequencing as a Window on the Epidemic | Dr. John Everett
References
- Andersen et al., 2020. The proximal origin of SARS-CoV-2. Nat Med. 26(4):450-452 https://pubmed.ncbi.nlm.nih.gov/32284615/
- Lau et al., 2020. Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 26(7):1542-1547. https://pubmed.ncbi.nlm.nih.gov/32315281/
- Papa et al., 2021. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLOS Pathogens. https://doi.org/10.1371/journal.ppat.1009246
- Petersen et al., 2020. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infectious Diseases. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30484-9/fulltext
- Wrobel et al., 202. Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2. Nature Communications, 12, 837. https://doi.org/10.1038/s41467-021-21006-9
- Wu et al, 2020. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 27(3):325-328 https://pubmed.ncbi.nlm.nih.gov/32035028/
- Wu & Zhao, 2021. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Research, 50. https://doi.org/10.1016/j.scr.2020.102115
Further Reading
Last Updated: Mar 10, 2021