Nov 15 2017
Researchers at Kanazawa University and the University of Tokyo report in Nature Communications the visualization of the dynamics of ‘molecular scissors’ — the main mechanism of the CRISPR-Cas9 genetic-engineering technique.
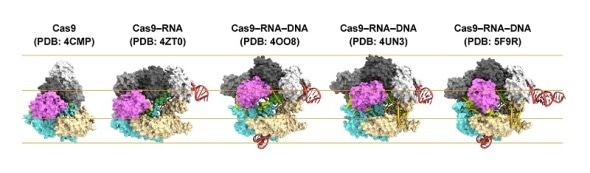
Structures of Cas9.
From left to right: Cas9 alone (apo-Cas9), Cas9 bound to RNA (Cas9–RNA), Cas9–RNA bound to its single-stranded DNA target (Cas9–RNA–DNA), Cas9–RNA bound to a partial DNA duplex (Cas9–RNA–DNA) and Cas9–RNA bound to its double-stranded DNA target (Cas9–RNA–DNA).
One of the techniques used in genetic engineering — the process of artificially modifying the genome of a living organism — involves the so-called CRISPR-Cas9 nuclease system. Using this system, a cell’s DNA can be cut at a desired site, where genes can be deleted or added. Selection of the site to be cut is done by a ‘guide RNA’ molecule bound to the Cas9 protein. Now, a team of researchers led by Mikihiro Shibata from Kanazawa University and Osamu Nureki from the University of Tokyo has visualized the dynamics of the CRISPR-Cas9 complex, in particular how it cuts DNA, providing valuable insights into the CRISPR-Cas9-mediated DNA cleavage mechanism.
For their visualization studies, the scientists used high-speed atomic-force microscopy (HS-AFM), a method for imaging surfaces. A surface is probed by moving a tiny cantilever over it; the force experienced by the probe can be converted into a height measure. A scan of the whole surface then results in a height map of the sample. The high-speed experimental set-up of Shibata and colleagues enabled extremely fast, repeated scans — convertible into movies — of the biomolecules taking part in the molecular scissoring action.
First, the scientists compared Cas9 without and with RNA attached (Cas9–RNA). They found that the former was able to flexibly adopt various conformations, while the latter has a fixed, two-lobe structure, highlighting the conformational-stabilization ability of the guide RNA. Then, Shibata and colleagues looked at how the stabilized Cas9–RNA complex targets DNA. They confirmed that it binds to a pre-selected protospacer adjacent motif (PAM) site in the DNA. A PAM is a short nucleotide sequence located next to the DNA’s target site, which is complementary to the guide RNA.
The research team’s high-speed movies further revealed that targeting (‘DNA interrogation’) is achieved through 3D diffusion of the Cas9–RNA complex. Finally, the researchers managed to visualize the dynamics of the cleavage process itself: they observed how the region of ‘molecular scissors’ undergoes conformational fluctuations after Cas9–RNA locally unwinds the double-stranded DNA.
The work of Shibata advances our understanding of the CRISPR-Cas9 genome-editing mechanism. In the words of the researchers:
… this study provides unprecedented details about the functional dynamics of CRISPR-Cas9, and highlights the potential of HS-AFM to elucidate the action mechanisms of RNA-guided effector nucleases from distinct CRISPR-Cas systems.