By looking into zinc-finger antiviral proteins (ZAP), a new study from Germany and the United Kingdom (UK) and available on the preprint server bioRxiv* suggests that zoonotic transmission of a coronavirus with a specific genetic make-up facilitated the pandemic spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), but also its sensitivity to interferons.
SARS-CoV-2, the causative agent of the ongoing coronavirus disease (COVID-19) pandemic, is spreading much more efficiently than the first zoonotic highly pathogenic coronavirus (SARS-CoV). But despite such rapid global spread, the virus seems to be more susceptible to inhibition by type I interferon when compared to its more severe variants (i.e., SARS and MERS).
.jpg)
Illustration of SARS-CoV-2, 2019 nCoV virusImage Credit: Orpheus FX / Shutterstock O By

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Interferons and zinc fingers
Interferon therapy induces the expression of hundreds of cellular interferon-stimulated genes, but it is currently unknown which of these genes contribute to inducible restriction of SARS-CoV-2 replication. Still, type I interferon is currently being considered in the treatment of COVID-19.
However, antiviral factors may exert strong selection pressure and give rise to specific viral properties that provide clues for efficient interferon-mediated immune responses. For example, coronaviruses are known to display marked suppression of CpG dinucleotides, and recent evidence points towards the conclusion that this is also the case for SARS-CoV-2.
Such CpG suppression may, at least partly, be driven by the ZAP that restricts manifold viral pathogens and specifically earmarks CpG-rich RNA sequences that are underrepresented in the human transcriptome (i.e., the set of all RNA transcripts).
In order to define selection pressures on SARS-CoV-2, but also other human coronaviruses, the researchers from Ulm University Medical Center in Germany and King's College London in the United Kingdom examined CpG frequencies and distribution in all seven human viruses and their closest animal counterparts.
For that purpose, a plethora of inventive techniques was employed: phylogenetic analyses, cell cultures, CpG content sliding window analysis, real-time reverse transcription-polymerase chain reaction (qRT-PCR), SDS-PAGE and Western blotting, plaque-forming Unit (PFU) assay, as well as TCID50 endpoint titration.
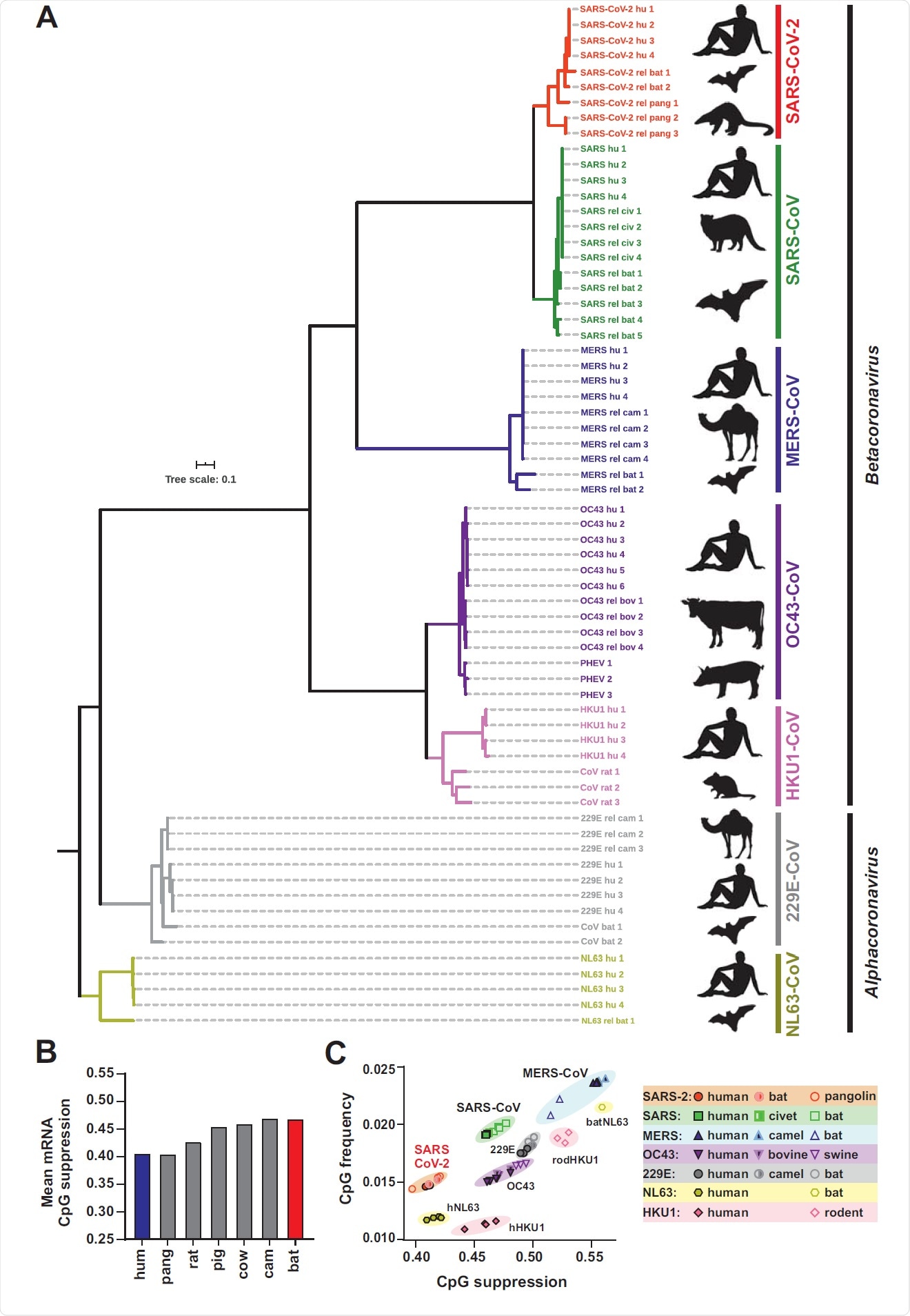
Phylogenetic relationship between human coronaviruses and their animal relatives and CpG suppression in pathogens and their hosts. (A) Distance-based relationship inference based on representative full genome nucleotide sequences of human SARS-CoV-2, SARS, MERS, HKU-1, OC43, NL63 and 229E strains and their closest known animal relatives. Black symbol to the right indicates the viral host (human, bat, pangolin, civet, camel, rat, pig, cattle). (B) Mean CpG suppression (i.e. number of observed CpGs normalized to expected CpGs based on sequence length and GC content) in mRNAs of the indicated host species (human, pangolin, rat, pig, cow, camel and bat). (C) CpG frequency (no. of CpGs normalized to sequence nucleotide length) and suppression (no. of observed CpGs normalized to expected CpGs based on sequence length and GC content) in human coronavirus genomes and their closest animal-infecting relatives. See also Table S1.
Different degrees of adaptations
The researchers have shown that ZAP – expressed in human lung cells together with its respective co-factors – specifically targets CpG dinucleotides in viral RNA sequences and can, therefore, restrict SARS-CoV-2. Moreover, interferon types I, II, and III strongly inhibited SARS-CoV-2 and further induced ZAP expression.
Both SARS-CoV-2 infection and interferon treatment-induced ZAP expression in cell-based systems used in the study. This finding agrees with the recent discovery that ZAP messenger RNA expression is clearly induced in SARS-CoV-2-infected human individuals.
Remarkably, SARS-CoV-2 and its closest bat relatives show the strongest CpG suppression among all known human and bat coronaviruses. Still, knock-down of ZAP significantly increased SARS-CoV production in lung cells, especially upon treatment with interferon-alpha or interferon-gamma.
"Consistent with an increased selection pressure for CpG suppression in the human host, we found that community-acquired human coronaviruses show lower frequencies of CpG dinucleotides than their closest animal relatives", explain study authors in their paper available on bioRxiv preprint server.
"This was not observed for highly pathogenic SARS and MERS coronaviruses, most likely reflecting less advanced human adaptation consistent with their less effective and transient spread," they add.
Altogether, the results support the notion that the selective pressure against CpGs is augmented upon zoonotic transmission from bats and most intermediate hosts to humans. This suggests that the differences between human coronaviruses and their animal relatives may reflect different degrees of adaptation.
Towards effective and safe immune approaches
"In summary, our results show that ZAP restricts SARS-CoV-2 and contributes to its inhibition by interferons", say study authors. "They further suggest that zoonotic transmission of coronaviruses already showing strong CpG suppression may have facilitated effective spread in humans," they add.
Their findings also highlight that SARS-CoV-2 is highly susceptible to interferons and might prompt the appraisal of combination therapies for the treatment of COVID-19, which includes interferon-gamma.
Nonetheless, we still lack a full understanding of viral immune evasion and counteraction mechanisms. While ZAP is an essential effector of the antiviral immune response, it is merely one of many that remain to be defined; hence, it will be pivotal to determine whether the different interferon types utilize different effectors to restrict SARS-CoV-2.
In any case, improved knowledge of antiviral effectors of the interferon response, as well as viral countermeasures, will show us a way forward to developing effective and safe immune therapy approaches against COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources