Novel Coronavirus SARS-CoV-2 Creative rendition of SARS-COV-2 virus particles. Note: not to scale. Credit: NIAID
The global coronavirus disease (COVID-19) pandemic is currently accelerating in different parts of the world, but so is the research on fundamental aspects of its causative agent, the virus named SARS-CoV-2. Nonetheless, we are still without an approved drug or vaccine, which means many lives are at stake.
What we know is that the SARS-CoV-2 nucleocapsid protein is a copious RNA binding protein that plays numerous roles in the viral life cycle, such as replication, transcription, and genome packaging. It shares many sequence features with other nucleocapsid proteins from coronaviruses.
A body of work on nucleocapsid protein from a plethora of model coronaviruses has revealed that it undergoes self-association, specific interaction with other proteins, as well as interaction with RNA nucleic acid – all in a highly multivalent manner.
However, despite its decisive and multifunctional nature, the molecular details on how nucleocapsid protein mediates these functions are still poorly understood. Only recently, it was reported that nucleocapsid protein undergoes phase separation with RNA.
And since it is known that this protein underlies viral packaging, one of the hypotheses is that phase separation may actually represent an inevitable epiphenomenon that reflects physical properties essential in a protein to drive the compaction of long RNA molecules.
This is why a research group from the Washington University School of Medicine and Stony Brook University decided to combine single-molecule spectroscopy with all-atom simulations to unveil the molecular details that capture the function of SARS-CoV-2 nucleocapsid protein.
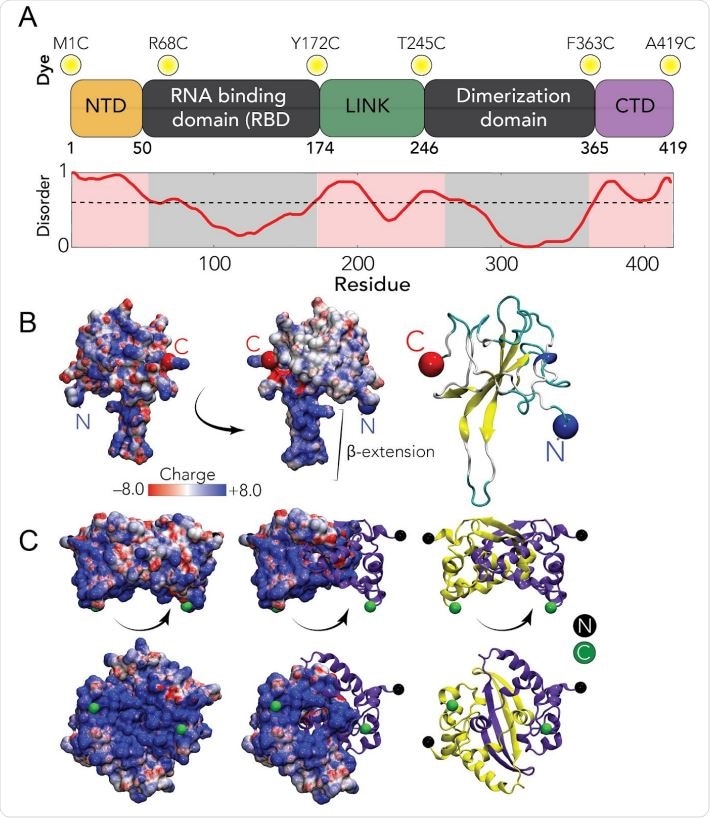
Sequence and structural summary of N protein A Domain architecture of the SARS-CoV-2 N protein. Dye positions used in this study are annotated across the top, disorder prediction calculated across the bottom. B Structure of the SARS-CoV-2 RNA binding domain (RBD) (PBD: 6yi3). Center and left: colored based on surface potential, revealing the highly basic surface of the RBD. Right: ribbon structure with N- and C-termini highlighted.
Full-length constructs of the nucleocapsid protein
The goal of this approach was actually to reconstruct an atomistic description of the nucleocapsid protein conformational ensemble, as well as to map its intrachain interactions. To this end, this research group created three full-length constructs of the nucleocapsid protein with fluorescent labels.
"These constructs allowed us to probe conformations and dynamics of the disordered regions in the context of the full-length protein using single-molecule Förster Resonance Energy Transfer (FRET) and Fluorescence Correlation Spectroscopy (FCS)," study authors explain their approach.
In parallel to these experiments, they have also performed all-atom Monte Carlo simulations of each of the three intrinsically disordered regions in isolation and in context with their adjoining folded domains.
Untangling the complex nexus of interactions
This study found that nucleocapsid protein firmly undergoes phase separation in vitro with model RNA under a range of different salt conditions. By utilizing simple polymer models, the researchers further revealed that the same interactions that drive phase separation might also drive genome packaging into a dynamic, single-genome condensate.
More specifically, this model suggested that the symmetry breaking through a small number of high-affinity binding sites can indeed organize anisotropic multivalent interactions to drive single-polymer compaction, quite dissimilar to multi-polymer phase separation.
"We propose a simple model in which symmetry breaking through specific binding sites promotes the formation of metastable single-RNA condensate, as opposed to large multi-RNA phase-separated droplets," said study authors
And as observed for other proteins with similar molecular features, the researchers have found that nucleocapsid protein undergoes liquid-liquid phase separation when mixed with RNA. Moreover, polymer models forecast that the same multivalent interactions that drive phase separation also engender RNA compaction.
"Irrespective of its physiological role, our results suggest that phase separation provides a macroscopic readout (visible droplets) of a nanoscopic process (protein: RNA and protein: protein interaction)," further clarify study authors.
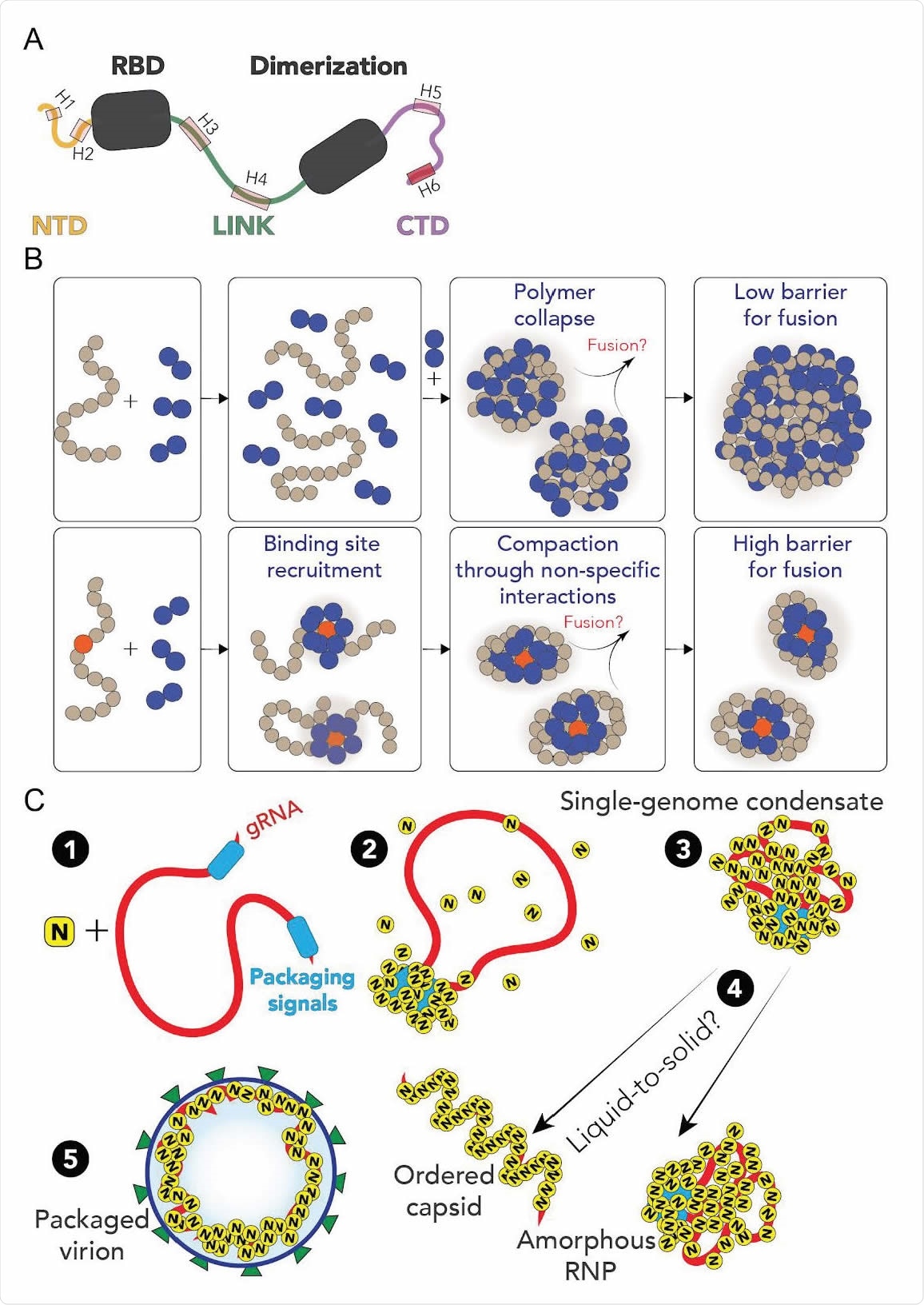
A. Summary of results from single-molecule spectroscopy experiments and all-atom simulations. All three predicted IDRs are disordered, highly flexible, and house a number of putative helical binding regions that overlap with subregions identified previously to drive N protein function. B. Overview of general symmetry breaking model. For homopolymeric polymers, local collapse leads to single-polymer condensates that undergo barrier-less fusion, rapidly assembling into large multi-polymer condensates. When one (or a small number of) high-affinity sites are present, local clustering of binders at a lower concentration organize the polymer such that single-polymer condensates are kinetically stable. C. Proposed model for SARS-CoV-2 genome packaging. (1) A simplified model of SARS-CoV-2 genome with a pair of packaging region at the 5’ and 3’ end of the genome (2) N protein preferentially binds to packaging signal regions in the genome, leading to a local cluster of N protein at the packaging signal RNA. (3) The high local concentration of N protein drives condensation of distal regions of the genome, forming a stable single-genome condensate. (4) Single-genome condensates may undergo subsequent maturation through a liquid-to-solid (crystallization) transition to form an ordered crystalline capsid or solidify into an amorphous ribonuclear particle (RNP). While in some viruses an ordered capsid clearly forms, we favor a model in which the SARS-CoV-2 capsid is an amorphous RNP. Compact single-genome condensate ultimately recruits E, S and M proteins at the membrane, driving envelope formation and final virion packaging.
Drugs that disrupt viral packaging
According to their results, the researchers postulate that viral packaging may involve initial genome compaction through multivalent protein: RNA and protein: protein interactions, which is then followed by a liquid-to-solid transition in cases where well-defined crystalline capsid structures emerge.
"We speculate that RNA compaction to form dynamic single-genome condensates may underlie the early stages of genome packaging," say study authors. "As such, assays that measure how compounds modulate phase separation could provide a convenient tool for identifying drugs that disrupt viral packaging," they add.
The model is also in sufficient empirical agreement with recent observations made for other viral agents, suggesting in turn that nature is leveraging the same principles as an evolved mechanism for monodisperse particle assemblage.
In conclusion, there are significant therapeutic implications of both understanding and modulating phase separation here (but also elsewhere in biology), and these results offer a window into critical physical interactions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Cubuk, J. et al. (2020). The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. bioRxiv. https://doi.org/10.1101/2020.06.17.158121.
- Peer reviewed and published scientific report.
Cubuk, Jasmine, Jhullian J. Alston, J. Jeremías Incicco, Sukrit Singh, Melissa D. Stuchell-Brereton, Michael D. Ward, Maxwell I. Zimmerman, et al. 2021. “The SARS-CoV-2 Nucleocapsid Protein Is Dynamic, Disordered, and Phase Separates with RNA.” Nature Communications 12 (1): 1936. https://doi.org/10.1038/s41467-021-21953-3. https://www.nature.com/articles/s41467-021-21953-3