The current COVID-19 pandemic caused by a single-stranded RNA virus, thought to have jumped across species barriers to infect humans, has spread rapidly across the globe infecting over 10 million individuals. The virus, now known as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the causative agent of COVID-19 disease. COVID-19 disease is a respiratory illness where symptoms can range from very mild to severe and include fever, coughing, a sore throat, and shortness of breath.
As of now, the search for antivirals and therapeutics is well underway, but the lack of a complete understanding of the viral structure and biology remains a challenge.
Now, a new study by scientists ar the Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology and the Boston University School of Medicine and published on the preprint server bioRxiv in June 2020 describes the RNA structures within the virus that play a critical regulatory role in viral transcription, and also reveal significant differences in the structure of some major drug targets within the virus.
Secondary RNA Structures Important for Replication and Transcription
The earlier studies on coronavirus structures show several conserved regions that play an essential role in viral replication, such as the 5’ UTR, the 3’ UTR, and the frameshift element (FSE). Their structure has also been computationally derived using experimental data, from techniques like RNAse probes and nuclear magnetic resonance (NMR) spectroscopy. Additionally, the functional importance of the secondary structures of these subgenomic regions has been reported for viral replication and transcription.
The Frameshift Element
The FSE, for instance, is near the ORF1a/ORF1b boundary and acts to frameshift the ribosomal reading frame by one nucleotide. This bypasses the stop codon at the end of the ORF1a and provides exposure for the ORF1b encoded proteins like the viral RNA-dependent RNA polymerase (RdRP). For many viruses, the rate of frameshift is strictly regulated as small changes in the percentage of frameshifting cause the production of genomic RNA and the viral dose required for infection to change dramatically.
For this reason, the FSE is a primary therapeutic target, and small molecule drugs are being investigated to alter the rate of this slippage at the ribosomes and thus inhibit the translation of viral proteins.

Genome-wide probing of intracellular SARS-CoV-2 RNA structure with DMS-MaPseq. (A) Schematic of the experimental protocol for probing viral RNA structures with DMS-MaPseq. (B) Correlation of DMS reactivities for each base between two biological replicates. (C) Genome-wide coverage as a function of position. Coverage at each position represents the average coverage over a 400 nt window. (D) Signal and noise as a function of genome position for untreated and DMS-treated RNA. Signal (mutation rate for A and C) and noise (mutation rate for G and U) at each position was plotted as the average of 100 nt window. Mutational Fraction of 0.01 at a given position represents 1% of reads having a mismatch or deletion at that position. (E) In-cell model of the 5’ UTR and beginning of ORF1a structure. Bases are colored by their DMS signal; bases that are not DMS reactive are colored white.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Absence of Validated FSE Model
The FSE of SARS-CoV is almost identical to that of SARS-CoV-2, with only one nucleotide difference. NMR study shows it to be a pseudoknot structure with 3 stems. The presence of the pseudoknot causes ribosomal pausing and then back-shifting by one nucleotide to relax the tension. As yet, however, the rate of such shifting in SARS-CoV-2 is unknown.
There is no validated RNA structure model for the FSE, and for many other functionally important genomic elements of CoVs are still unknown. This includes most of the transcription-regulating sequences (TRS). These short sequences are essential for the transcription of subgenomic RNA (sgRNAs), the basic transcribed sequence from which all the proteins that are encoded outside the ORF1ab are translated.
The genome of SARS-CoV-2 has 10 TRS, one located at the 5’ end, a leader TRS, and the rest body TRS, one before each of the open reading frames other than ORF1ab. These body TRS generate 9 sgRNAs by discontinuous transcription.
Early in silico computational models were improved using chemical probes for a genome-wide RNA analysis, since these provided essential in vivo measurements of the structures in vivo.
The Study: DMS Probing of RNA Structure
The current study uses the probe dimethyl sulfate (DMS) to probe the secondary structure of the whole RNA genome. The researchers say, “Our results reveal major differences within silico predictions and highlight the physiological structures of known functional elements. Our work provides experimental data on the structural biology of RNA viruses and will inform efforts in the development of RNA-based diagnostics and therapeutics for SARS-CoV-2.”
The researchers added DMS to infected Vero cells and carried out mutational profiling with sequencing since DMS rapidly pairs to unpaired adenines and cytosines in vivo. They found five stem-loops (SL) within the 5’ UTR and three stem-loops near the beginning of ORF1a. These have different vital functions.
Stem Loops Functionally Important
SL1 is needed for viral replication, SL2, which is the most highly conserved in this region, is required for replication, and SL3 contains the leader TRS needed for discontinuous transcription. SL4 is required for sgRNA synthesis, maintaining proper spacing between upstream and downstream stem-loops.
Structured and Accessible Regions
All stretches which had at least 10 consecutive paired or 14 unpaired bases are called structured or accessible, respectively. The former might be therapeutic targets, and there were 215 identified in this study, including some structures with known and many with unknown functions.
Accessible regions could serve as targets of antisense oligonucleotide therapies, and 261 of them were identified here. Importantly, 11 accessible regions are present in ORF-N, which is present in almost all sgRNAs, which means they may all be targeted by this type of therapy, together or individually.
They also found that TRS are located within stem-loops in 7/9 cases.
Alternative FSE Structure
The researchers also examined the FSE structure within cells and found that instead of the pseudoknot formation downstream of a slippery sequence where the transcript shifts one nucleotide, there is an alternative pairing achieved by half of the canonical stem 1, to 10 bases upstream, of the slippery sequence. This is called the alternative stem 1 (AS!).
This refolded structure is visualized only when the whole length of the RNA molecule is provided as the context of the FSE sequence. This is because only the whole-length genome provides the full range of pairing possibilities, which drives the adoption of the alternative stem 1, as the predominant structure. This pairing sequence is conserved across the sarbecovirus clade and seems to be unique to them.
The FSE also forms two different structures within the cells, structures 1 and 2, but further studies will be needed to find whether these contribute to frameshifting efficiency by stopping the ribosome at the slippery site rather than far away from it.
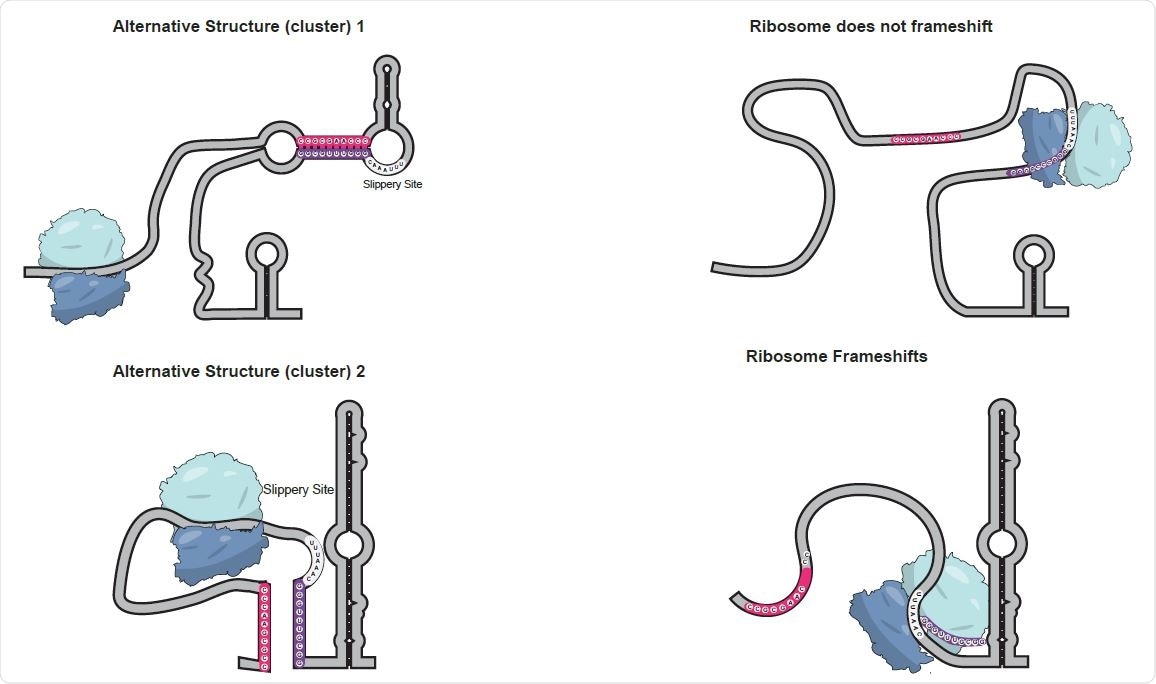
Model of alternative structures regulating frameshifting. When genomic RNA folds into Alternative Structure 1 (top), the slippery site resides within a loop in the middle of a long stem-loop. As the ribosome starts to unwind the RNA, it may pause at the base of the stem, but this pause is far from the slippery site. By the time the ribosome reaches the slippery site, the structure in front of it has been unwound. As the ribosome continues it will reach the upstream stop codon and terminate translation. In contrast, Alternative Structure 2 (bottom) forms a 75 nt stem loop right in front of the slippery site. This stem loop can cause the ribosome to pause, frameshift -1 nt and bypass the upstream stop codon to continue translation.
Implications and Therapeutic Importance
Previous work shows that the formation of pseudoknots or stable stems just upstream to the slippery sequence improves frameshifting rates. Still, no specific studies on translation efficiency of ORF1ab in sarbecovirus-infected cells are known.
The study also suggests the involvement of N protein in binding to and unwinding the TRS to allow it to function. If the stability of the TRS is altered, this could reduce its affinity for the N protein, which in turn could alter the expression of sgRNAs. This could, therefore, be another therapeutic target.
Overall, the study uncovers several major RNA structures over the whole genome and suggests a new FSE structural model with the primary model being alternative stem 1. The researchers point to the direction of new work: “Future work will involve determining by what mechanism and to what extent the alternative structures of the SARS-CoV-2 FSE regulate translation of ORF1ab, as well as whether the FSE can fold into a pseudoknot in cells.”
This type of understanding could allow the development of more precisely designed and effective therapeutics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources