.jpg)
Colorized scanning electron micrograph of a cell heavily infected with SARS-CoV-2 virus particles (yellow), isolated from a patient sample. The black area in the image is extracellular space between the cells. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
Although in January 2020 coronavirus disease (COVID-19) was still not a pandemic threat we are currently facing; the first SARS-CoV-2 genome sequence was already released, allowing, in turn, the development of tests that appraise the presence of viral RNA in biological samples. This quickly provided a way to identify individuals who are currently infected by the virus.
There is no consensus on how infectious asymptomatic people are. Still, there is no doubt that many individuals can transmit the virus before they develop symptoms or if they have a mild clinical presentation. Accordingly, routine screening of large parts of the population as the pandemic spreads becomes a necessity.
A trifecta of methods
LAMP is a well-established technique using targeted isothermal amplification that generates micrograms of product from tens of copies of the target region – all within 30 minutes at 65 °C. However, the precision is hampered by proxy measurement (i.e., fluorescent color changes or increased turbidity).
Conversely, sequencing techniques provide precise results, as well as an opportunity to amplify and detect multiple targets in a single reaction. During nanopore sequencing, an electrical current is calculated as template strands pass through each pore on the flow cell array – enabling data analysis in real-time.
Oxford Nanopore Technologies' (ONT) barcoded rapid library prep kit employs a transposase to convert DNA to a barcoded library that is amenable for sequencing in approximately 15 minutes, with 96 barcodes available. This allows prepared samples to be pooled for sequencing. Can all of these techniques be combined to increase sensitivity and specificity?
In this manuscript, the researchers from Oxford Nanopore Technologies demonstrated a method that indeed combines the rapid target-specific amplification provided by LAMP, a method of transposase-based library preparation, and real-time nanopore sequencing and data analysis. This method is also known as the triplex SARS-CoV-2 assay or LamPORE.
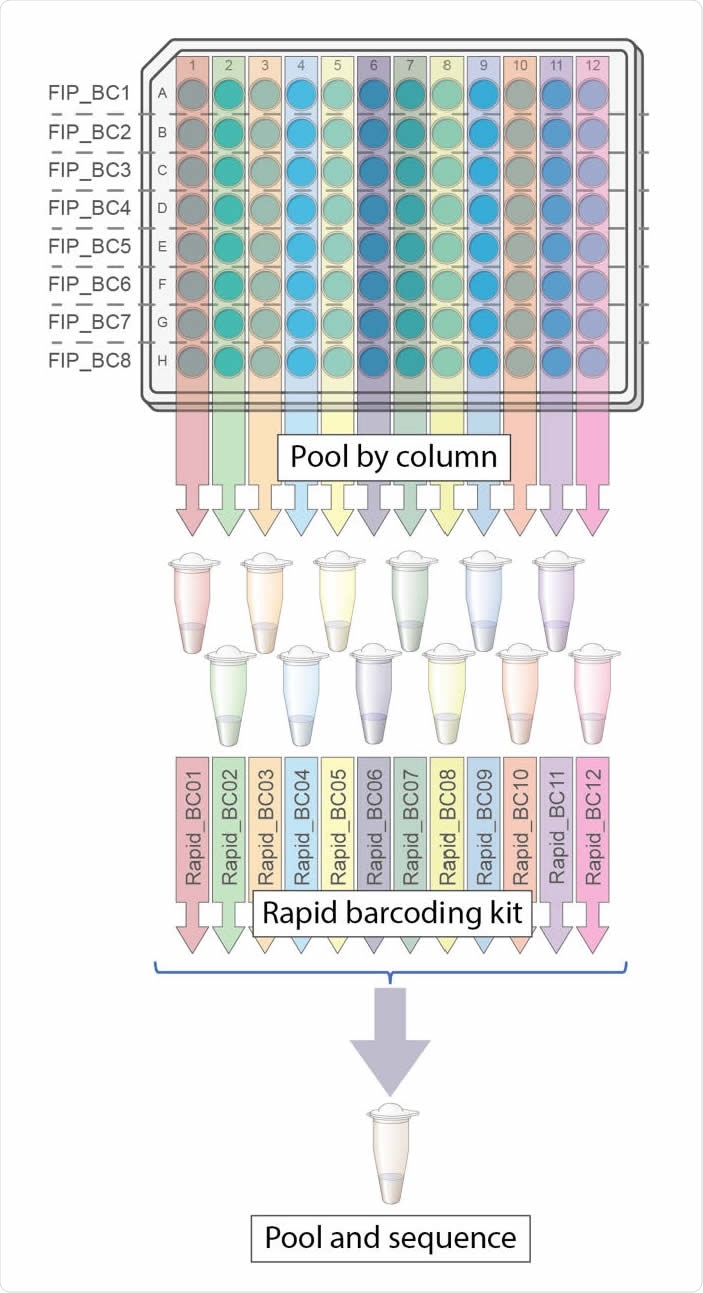
Overview of LamPORE laboratory workflow
Optimizing assay performance
To prepare the LamPORE assay, the researchers chose to target three different regions in the SARS-CoV-2 genome in a single multiplexed reaction. These are ORF1a, the envelope (E), and nucleocapsid (N) genes, with primer sets AS1, E1, and N2, respectively.
As a control for the quality of the sample preparation, RNA extraction, reverse transcription, and LAMP amplification, the scientists included a set of primers in order to amplify the human actin messenger RNA. The latter should be present in all swab samples (irrespective of their SARS-CoV-2 status); therefore, this can differentiate between true negatives and invalid samples.
Furthermore, to appraise the inclusivity of the proposed triplex SARS-CoV-2 assay, they have aligned all primer sequences to the 46,872 human SARS-CoV-2 genomes deposited at GISAID (as of June 16, 2020). The presence of a single mismatch will not likely influence the limit of detection.
Receiver operating characteristic (ROC) curves were generated for the SARS-CoV-2 read count sum, and also for read counts from each individual SARS-CoV-2 target. The sum of read counts across each of the three SARS-CoV-2 targets (AS1, E1, and N2) served as the scoring metric for deciding whether the results are positive, negative, inconclusive, or invalid.
To expand the evaluation of the assay's performance, the researchers have obtained eighty clinical samples positive for SARS-CoV-2 RNA by RT-qPCR, which had been extracted from nasopharyngeal swabs of infected individuals.
Quick and accurate results with high scalability
In this study, the researchers have shown that a multiplexed amplification reaction where three separate regions of the SARS-CoV-2 genome are targeted translates to high sensitivity performance, highly comparable with reverse transcription-polymerase chain reaction (RT-qPCR).
Starting with extracted RNA, the results can be obtained from 12 samples in approximately an hour, and from 96 samples in less than 2 hours. Moreover, high scalability is readily achieved by combinatorial barcoding, and template quantities ranged from 20 to 250 copies per reaction.
Out of the 80 RT-qPCR-verified positives, 79 were called positive in the LamPORE analysis. The false negative corresponded to the sample with the lowest viral load (according to the cycle quantification value obtained with PCR).
Greater degrees of multiplexing
In short, this novel assay is simple to scale from small sample numbers to thousands, with greater degrees of multiplexing that is feasibly achieved by increasing the numbers of barcodes, as well as with the use of combinatorial barcoding.
"LamPORE relies on sequencing, as opposed to a color change, which raises the possibility of using a single multiplexed LamPORE reaction to detect many different pathogens," highlight study authors in their medRxiv paper.
When co-infections are concerned, this assay should also identify the exact combination of pathogens present in the sample. Performance characteristics are currently being established, and regulatory clearance to market is underway, which is encouraging news in our fight against COVID-19 pandemic.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
James, P. et al. (2020). LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv. https://doi.org/10.1101/2020.08.07.20161737.