As the COVID-19 pandemic continues, scientists continue to scout new methods to predict or detect early changes in the prevalence of the infection in order to help evolve more effective public health strategies and contain its spread. A new study published on the preprint server medRxiv* in September 2020 describes the measurement of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins in wastewater as a reliable way to track the virus in the community.
SARS-CoV-2 RNA in wastewater has recently gained ground as a method of understanding the prevalence of COVID-19 in a community, by using polymerase chain reaction (PCR) to amplify the nucleic acid. However, the low levels of fecal viral RNA, and the instability of RNA, compounded by a warm environment in many cases, as well as exposure to harsh chemicals and conditions, mean that cycle thresholds above the limit of detection of the technology are often required to amplify it to detectable levels.
Another shortcoming of this technology is that the sewage preparation needs extensive processing, making it more expensive and cumbersome.
Using MPAD for Viral Protein Detection in Wastewater
To overcome these obstacles, researchers have been examining the feasibility of measuring viral proteins instead, since these are present at a higher load and are more stable. The current study uses the MPAD (Multiplex Paired-antibody Amplified Detection), which is remarkable for coupling the specificity of antibodies with the signal amplification capability of PCR. The aim was to evaluate the ability of this method to signal the presence of the virus more strongly and reliably, using wastewater samples derived from two Ottawa wastewater recovery facilities.
Ottawa, Canada, has a low prevalence of COVID-19, and less than 10 cases are reported daily per 100,000 population. Nonetheless, the same team of researchers had earlier discovered detectable levels of viral protein in sewage samples. They chose this city as it would provide a useful standard for assay sensitivity in terms of viral detection. They also determined to use daily hospital admissions as a measure of viral shedding in the community, in preference to positive tests, since the former is relatively unchanging and more accurate.
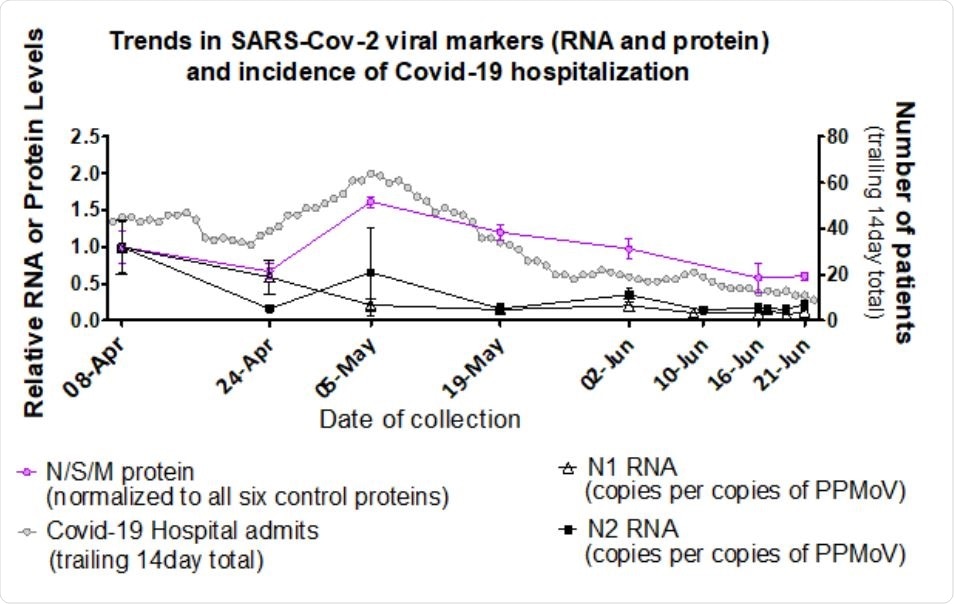
Levels of Ottawa wastewater SARS-CoV-2 structural proteins and RNA correspond with COVID-19 hospitalizations. Pink circles (from Figure 5): left Y-axis; MPAD determined Ottawa PEG precipitated influent solids N, S and M protein levels relative to April 8, 2020 (set to a value of 1.0), normalized to geomean of six fecal control proteins (see methods). Collection dates specified on the X-axis (no sample was analysed for June 10). Error bars represent standard deviation between replicates. Black triangles and squares (From D’Aoust et al., 2020)8 : left Y-axis; RT-PCR generated Ottawa PEG precipitated primary sludge viral RNA (N1 hollow triangle and N2 solid square) relative to level on April 8 (set to a value of 1.0) normalized to PMMoV. Collection dates specified on the X-axis, including additional samples on June 17 and 19. Grey circles: right Y-axis, 14-day (estimated median duration of fecal viral shedding) trailing sum of daily hospital admissions, proxy for active viral shedders in the community.
Maximizing the MPAD Advantage
MPAD uses quantitative PCR, just like the qRT-PCR used for viral RNA detection. This means the cycle thresholds can be directly compared, as well as the signal for protein detection vs. RNA detection. MPAD also allows several viral proteins to be analyzed simultaneously to increase the specificity of detection.
Wastewater is a highly variable system with regards to flow, mass, type and concentration of organisms, and other vital characteristics. The RNA stability also varies with the conditions. The researchers used a multiple marker technique to standardize the wastewater sample in terms of fecal content.
They, therefore, combined a surrogate fecal virus, PMMoV, which was earlier shown to be the most reliable of three different approaches, and five other content markers, including two viruses and three human proteins, with the three SARS-CoV-2 proteins to be assayed. These content markers allowed each of the three proteins to be normalized to the six fecal content markers, both separately and in combination.
They also checked the correlation between the markers for each protein, comparing the levels normalized to the individual controls vs aggregated controls. This confirmed a highly superior normalization to be achieved with this combination of markers, with the best accuracy and comparability.
The researchers remark, “The robust specificity and normalization devolving from measuring all 9 (3 SAR-CoV-2, 6 fecal control markers) protein levels with this single, multiplex protein panel represents a significant advantage in any broad wastewater surveillance program.”
Markedly Higher Detection Rates in International Samples
When processed by MPAD quantitative assay, it was found that the signal was several orders of magnitude higher than for RNA using qRT-PCR. In primary sludge, the viral protein signal was 64 to 208 times higher, while with influent solids, it was 20-128 times more significant than for viral RNA in the same samples.
However, these signals come from several cities with a wide range of prevalence. Once adjusted, the difference in signal between protein and RNA ranges from 400 to 6,500 times higher in primary sludge, and 100-1,000-fold in influent solids.
The signal correlated with the RNA and also the estimated incidence of the infection over the 10 weeks of the study period.
Testing Ottawa Samples
With the Ottawa samples, the researchers first did Western blot analyses to look for the presence of the virus in wastewater. Their findings confirmed those of Raman spectroscopy, with spike protein being present in all samples of a series taken over time, unlike viral RNA, which was present only in the most upstream specimens. The quantity of viral membrane protein detected agreed with the trend of viral RNA.
They then measured the proteins using MPAD as part of the expanded panel. They found that they were able to detect viral RNA in influent solids on four of seven dates, but protein in all samples using MPAD. This could be because the protein content is higher because MPAD is more sensitive than qRT-PCR or both factors.
Secondly, the viral RNA in influent solids did not correlate with that in primary sludge, with the viral protein signal, or with clinical data. They concluded that the viral RNA signal in influent solids probably was at the limit of detection of the qRT-PCR and would not be a good indicator of the viral titer in wastewater, compared to primary sludge.
In the latter, the MPAD viral protein analysis produced a signal that matched the corresponding temporal trend observed in influent solids, and also the rolling 14-day total of hospital admissions for COVID-19.
Implications and Future Directions
The researchers concluded that it is possible to detect and measure the structural proteins of the virus in primary sludge after PEG precipitation. The data also suggest the presence of an intact virus, though the infectivity of sewage was not confirmed. Finally, this panel was able to control for the high variability of wastewater-based prevalence monitoring.
This finding, they say, will help study the epidemiology of the disease, being more sensitive to the presence of the virus than RNA detection-based methods. Also, by providing early warning, it can help track the viral load in the community as a relatively simple method of surveillance, whether for institutions, schools, or homes.
The measurements of both viral RNA and protein in stool in relation to the clinical activity of the virus suggests that the virus is shed in stool for about 14 days (median value). This would suggest that the rolling total of daily admissions for the last 14 days before a specified day indicate the number of hospitalized people who are shedding virus on any given date.
However, a more extended period of study is required to understand how well viral protein in wastewater is concordant to the prevalence. More frequent sampling over this period will show the changing temporal trends of community prevalence. Nonetheless, the study concludes, “The protein viral metrics in this study tracked well with this limited estimate of community viral load and with a sensitivity greatly exceeding that of viral RNA tracking.’
Source

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.