After screening more than 3,500 FDA-approved drugs and small molecules, researchers at the Chinese Academy of Sciences found that ceftazidime, an antibiotic approved for use in treating pneumonia, binds to the receptors that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) binds to in human cells. This drug could potentially be used to prevent virus infection. The research is published on the bioRxiv* preprint server in September 2020.
To combat the spread of COVID-19, researchers worldwide are racing to find strategies to prevent the SARS-CoV-2 coronavirus from infecting more people. Some methods by which the virus attacks human cells are now known, and these have guided treatment approaches.
The coronavirus has proteins, called spike proteins, which jut out from its surface. A portion of the proteins has areas called receptor binding domains (RBDs) that can bind to receptors of the angiotensin-converting enzyme 2 (ACE2) found on the host cells. Once the RBDs bind to the ACE2 receptors, the virus can fuse with the host cell membrane and infect the cell.
Several strategies for fighting the virus infection rely on preventing the virus from binding to the host cell receptors. For example, many monoclonal antibodies have been developed that target the spike protein of the virus.
However, these strategies cannot be deployed immediately as they need to undergo large scale trials for efficacy and safety in humans, which have long timelines. Furthermore, the cost of monoclonal antibodies is very high, which may prevent its widespread use.
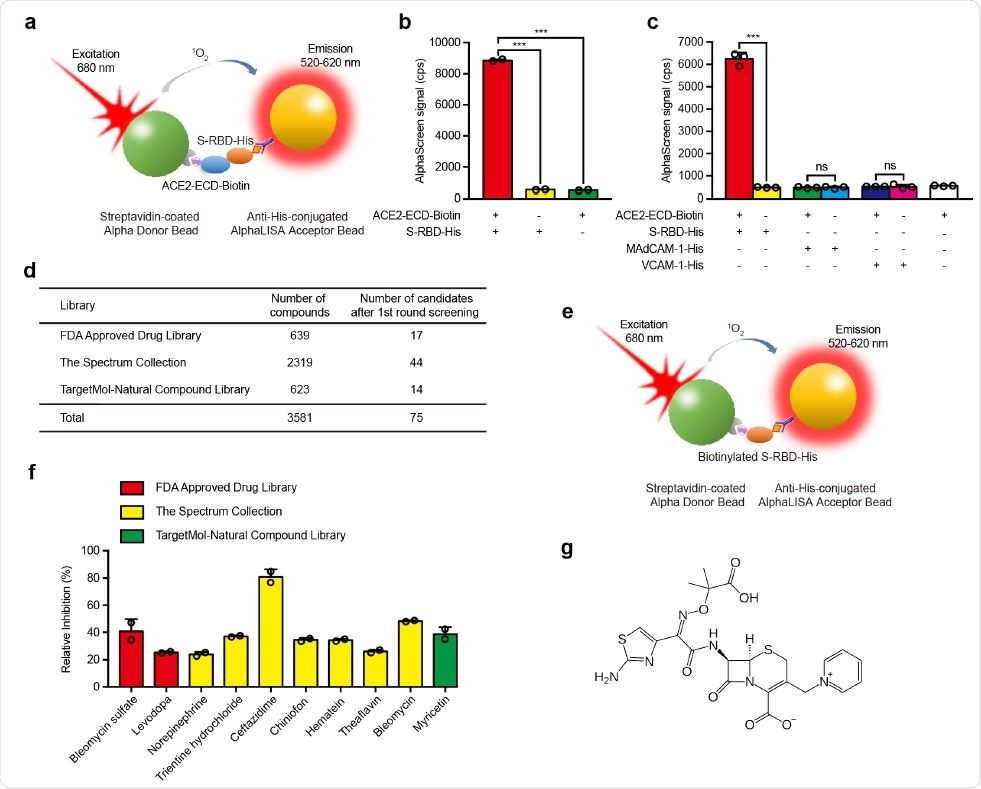
Screening of small molecule compounds that specifically block the interaction between S-RBD and ACE2. a, Schematic diagram of AlphaScreen system to detect the interaction between S-RBD and ACE2-ECD. The donor and acceptor beads are coated with streptavidin and anti-His monoclonal antibody, respectively. b, The interaction between S-RBD and ACE2-ECD was monitored using AlphaScreen 118 system. c, Comparison of the AlphaScreen signal of S-RBD-His, MAdCAM-1-His and VCAM-1-His proteins in the presence or not of ACE2-ECD-Biotin in AlphaScreen system. d, Libraries used in AlphaScreen-based high-throughput system, and 75 candidates were identified from 3581 compounds in positive selection. The inhibition rate was calculated by the decrease of AlphaScreen signal of each compound compared with that of DMSO vehicle control group. e, Schematic diagram of negative selection using AlphaScreen system. Biotinylated S125 -His simultaneously links streptavidin-coated donor bead and anti-His-conjugated acceptor bead together to generate AlphaScreen signal directly. f, Relative inhibition of 10 candidate compounds on S-RBD–ACE2 interaction using AlphaScreen system. The relative inhibition rate was calculated by subtracting the inhibition rate in negative selection from that in positive selection. g, Molecular structure of ceftazidime.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Repurposing known drugs
Another strategy to combat the virus is to figure out if drugs already approved for other diseases may work against SARS-CoV-2. Since these drugs are already approved for use in humans, their use can be almost immediate after proving that they do indeed work in preventing COVID-19.
Using this idea, researchers screened 3,581 small molecules from the FDA Approved Drug Library, Spectrum Collection, and Targetmol Natural Compound Library using a high throughput screening method, AlphaScreen.
The team purified RBDs and attached them to acceptor beads and the purified extracellular part of ACE2 to donor beads. When the RBDs bind to ACE2, the acceptor beads come close to the donor beads. Upon shining light of 680 nm wavelength, there is emission in the 520–620 nm region. A higher signal indicates greater affinity. If the small molecules screened prevent binding of the donor and acceptor beads, or lower signal, this indicates successful inhibition of binding of the RBDs to the ACE2 receptors.
The team identified 75 compounds after the first round of screening, showing at least 45% inhibition. They further refined their method to select 10 compounds from these molecules, which included bleomycin sulfate, levodopa, norepinephrine, trientine hydrochloride, ceftazidime, chiniofon, hematein, theaflavin, bleomycin, and myricetin.
Ceftazidime shows promise
Of these 10, ceftazidime was the most potent, with a relative inhibition rate of about 81%.
Ceftazidime is a type of cephalosporin antibiotic. It is an antibiotic generally used to treat bacterial pneumonia and is considered safe except for persons with allergic reactions to cephalosporins.
Upon further testing ceftazidime, the researchers found that it binds specifically and firmly to the spike proteins RBDs, but not the ACE2 extracellular domain. Ceftazidime also prevented binding of the coronavirus RBDs to the human pulmonary alveolar epithelial cells or lung cell, that also express ACE2.
Furthermore, ceftazidime also prevented the entry of the SARS-CoV-2 pseudovirus, an alternative to the natural virus with similar infectivity, into cells expressing the human ACE2. The concentration of ceftazidime required for 50% inhibition, or the IC50, was about 113 µM, and it did not show any measurable toxicity up to 400 µM.
In the treatment of pneumonia, the ceftazidime blood concentration can reach 300 µM. The authors say that at this concentration, ceftazidime showed 96% inhibition of SARS-CoV-2 pseudovirus infection in vitro.
Apart from ceftazidime, the researchers also tested 14 other cephalosporins, such as cephradine and cefazolin. However, only ceftazidime inhibited the binding of SARS-CoV-2 to ACE2.
The authors think that the action of ceftazidime is because of the chemical functionalities 2-aminothiazole, oxime with a terminal-exposed isobutyric acid, and the positively charged pyridine. These moieties could be interacting with the spike protein RBDs, preventing binding on the virus to the ACE2 receptors. The strong binding of ceftazidime to the RBDs suggests covalent bonding.
Because of the promising inhibition of ceftazidime to the virus, the authors suggest this drug should be considered for the treatment of COVID-19, especially as it is already approved, is safe, and affordable. Also, modifications of the chemical structure of the molecule could also be investigated to improve its inhibitory action further.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lin C. D. et al. (2020) Ceftazidime Is a Potential Drug to Inhibit SARS-CoV-2 Infection In Vitro by Blocking Spike Protein-ACE2 Interaction. bioRvix. https://doi.org/10.1101/2020.09.14.295956, https://www.biorxiv.org/content/10.1101/2020.09.14.295956v1
- Peer reviewed and published scientific report.
Lin, ChangDong, Yue Li, YueBin Zhang, ZhaoYuan Liu, Xia Mu, Chenjian Gu, Jing Liu, Yutang Li, GuoHui Li, and JianFeng Chen. 2021. “Ceftazidime Is a Potential Drug to Inhibit SARS-CoV-2 Infection in Vitro by Blocking Spike Protein–ACE2 Interaction.” Signal Transduction and Targeted Therapy 6 (1): 1–4. https://doi.org/10.1038/s41392-021-00619-y. https://www.nature.com/articles/s41392-021-00619-y.