Currently, researchers are studying the characteristics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as well as the host immune responses, in order to come up with better options for preventive and therapeutic approaches to containing this pandemic.
A new study by the Argentinian AntiCovid Consortium and published in the preprint server bioRxiv* in September 2020 reports on the successful characterization of the conformation and stability of the SARS-CoV-2 spike protein receptor-binding domain (RBD) in two different cell systems. This may help to accelerate the development of vaccines and neutralizing antibodies.
RBD Expression in Mammalian and Eukaryotic Cells
The RBD contains 220 amino acid residues with two N-glycosylation sites. N-glycosylation may be vital for protein folding, stability, and solubility, as well as on internal mobility and immunogenicity. The protein is a non-globular protein.
The researchers expressed the RBD protein in two different cell systems, allowing them to understand more about its stability and conformation. They first confirmed that this protein could not be expressed in E. coli, a cheap way to produce many other proteins, since it contains four disulfide bonds and is not spherical in shape. Instead, it yielded insoluble inclusion bodies which could not be rendered soluble by fusing it with various proteins. However, when tagged with glutathione S -transferase (GST), it became soluble, but because of its strong binding to the bacterial chaperone GroEL, it was not suitable for further applications.
They then proceeded to express it in the commonly used mammalian cell line HEK-293T and the yeast P. pastoris cell line. P. pastoris has several advantages. It is cheap and uses methanol as a carbon source as well as for the induction of the AOX1 promoter that drives the expression of the recombinant protein. It reaches high cell densities in bioreactor conditions. It also secretes the recombinant protein efficiently into the culture medium without high levels of endogenous proteins, simplifying the process of purification of the recombinant secretory protein.
The researchers found that in both cell lines, RBD expression led to the production of significant amounts of the soluble polypeptide demonstrating proper folding. When tested by fluorescence assays, their structure showed high similarities with the RBD protein of SARS-CoV expressed in P. pastoris.
They characterized both RBD expressions in the two cell types, using chemical and mass spectrometry analyses. They found that the latter failed to readily detect one specific peptide, probably because it is glycosylated at two positions, N331 (NIT) and N343 (NAT). This leads to an increase in mass, which removes it from the spectrum of MS mass analysis. Again, another peptide in the HEK-293T-expressed RBD was poorly detected, perhaps due to O-glycosylations in this sequence.
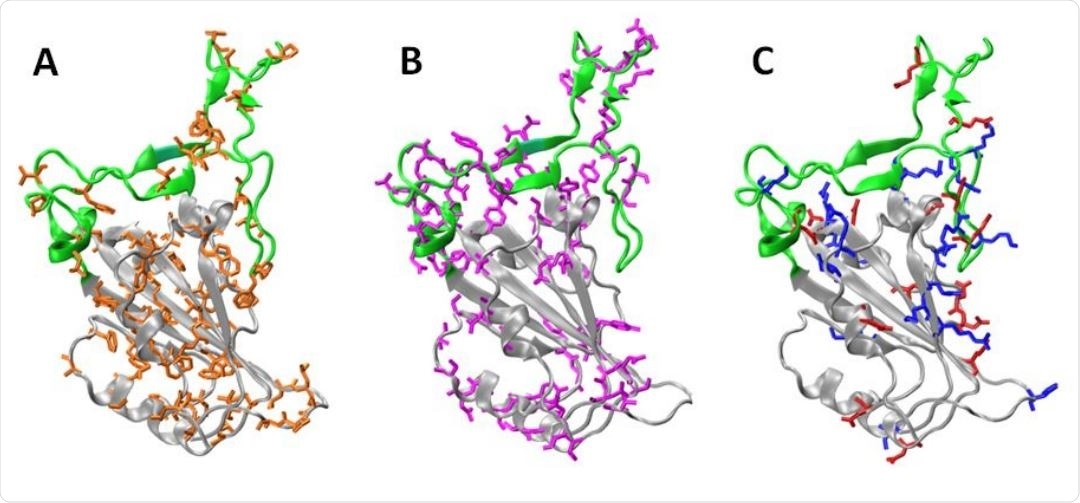
Subdomains and distribution of residue types on RBD . The Core (gray) and the RBM (green) regions are shown. Panels A, B, and C, shows the non-polar residues (orange: A, C, G, I, L, M, F, P, W and V), polar (violet: N, Q, S, T and Y), and charged residues (blue: basic K, R and H, red: acid D and E), respectively. To build the models we used the chain E of pdb structure 6m0j.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
N-Glycosylation Important for Proper Folding
It is known that proteins trafficked to the endoplasmic reticulum (ER) undergo N-glycosylation along with the translation, and the same glycan Glc 3 Man 9 GlcNAc 2 is transferred to the consensus sequence NXS/T in proteins in mammalian, plant, and yeast cells. In the ER this glycan is immediately removed, since they are important both for solubility and to control the protein folding.
Repeated cycles of glycan addition and removal in the ER continue until folding of the glycoprotein is completed. After that, the protein resumes its transport through the secretory pathway. Improperly folded pathways are transported back to the cytoplasm instead and broken down by protease enzymes inside the proteasomes.
Species-specific Glycosylation
The properly folded glycoproteins leaving the ER undergo changes involving the N-glycans in the Golgi apparatus so that finally they bear species-specific glycans. This includes mannose-rich yeast N-glycans on mature proteins, but complex or hybrid glycans in mammalian cell proteins.
The researchers found that when these two types of sugars were removed from the RBD expressed by both P. pastoris and HEK-293T cells, a similar band of ~26 KDa was present in both cases, corresponding to the molecular weight of the non-N-glycosylated RBD. If another treatment was applied to remove mannose-rich glycans only, the same band was obtained for the P. pastoris-expressed RBD but not that from the HEK-293T cells, supporting the complex or hybrid nature of glycans on this variant.
Removal of all the N-glycosylated RBD from the spike protein expressed in HEK-293T cells resulted in two protein forms, probably because of two O-glycosylated isoforms or two peptide isoforms. Earlier research has shown that O-glycosylation occurs at T323 and S325, supporting this possibility.
Glycosylation Does Not Affect Structure of Polypeptide
Heavy glycosylation of yeast secretory recombinant proteins with high-mannose glycans could impact the function of the proteins. Fortunately, the researchers did not find evidence that the different glycosylation patterns in the two types of cells affected the polypeptide structure in terms of either stability or conformation.
RBD is Stable Over pH and Concentration Variations
For both forms, the stability declined to a similar extent when the ionic strength was increased. Both were sensitive to changes in the ionic strength to the same degree. This indicates that ionic interactions lend stability to the structure of the RBD, and probably also regulate the internal mobility of this protein as well. Unfolding of the RBD in a medium without a reducing agent was reversible. Temperature-induced unfolding occurred at similar temperatures for both RBD types.
The mutated version of the SARS-CoV RBD had a significantly higher melting temperature compared to the wildtype SARS-CoV-2 RBD, perhaps because of differences in the ionic interactions or because this mutation suppresses N-terminal glycosylation. A third possibility is that the former simply has a more stable build.
The study concluded that native RBD remained stable at a range of pH and concentrations, even when frozen and thawed, without aggregating. This lends its stability under storage conditions without the need for stabilizers like glycerol. Thawing was occasionally followed by RBD precipitation when the protein concentration was 80 μm.
P. pastoris RBD is Scalable and Immunogenic
The RBD from P. pastoris elicited mouse antibody that detected RBD expressed from both cell types. The P. pastoris expression was also scalable, and could be carried out in a bioreactor to produce more than 45 mg/L of protein at over 90% purity.
This is suitable for producing large amounts of the protein cheaply. The researchers remark, “It is of high importance to produce and purify RBD at low-cost and efficiently, since this domain is extensively used for the development of serological test kits as well as an immunogen, both for the production of animal immune sera and for vaccine development.”
Implications and Future Directions
The researchers say their work helped to achieve some objectives of the SARS-CoV-2 consortium, namely, to study the RBD in detail and to come up with an inexpensive and high-yield RBD production system. They found that “RBD obtained from both yeast and mammalian cells were properly folded. They also had similar stability.”
They suggest that if the Sortase-A enzyme recognition site is incorporated into the RBD gene sequence. This would allow the RBD to couple covalently with fluorescent probes and peptides as well as with inexpensive protein carriers that can be churned out at low cost via E. coli and other cell systems, via transpeptidation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources