As the search for effective antivirals against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues, a new study published on the preprint bioRxiv* in November 2020 discusses the role of synthetic nanobodies or sybodies in neutralizing the virus by binding to its receptor-binding domain (RBD) on the spike protein.
Despite lockdown measures and social distancing, it remains difficult to control the spread of this virus. This is because of its high transmissibility, the absence of herd immunity, and the significant percentage of asymptomatic transmission. Many cases end in severe morbidity or death; over 1.2 million individuals worldwide have already lost their lives. It is thus imperative that we rapidly develop effective treatments to help mitigate the impact of those infected. This could help patients recover and prevent viral spread if used to treat the asymptomatic infection before they infect further transmission occurs.
The researchers focused on developing a candidate vaccine in the form of a monoclonal antibody, in view of the specific and potent nature of such treatments. They chose to raise the antibody against the full prefusion spike protein rather than against the purified RBD because previous studies have shown that antibodies raised against the latter do not show high neutralizing capacity, as expected.
The researchers used nanobodies (which are single-domain antibodies containing the variable VHH domain of camelid antibodies) possessing only the Ig heavy chains. These are found to have immense potential, being small, stable and cheaply produced in bacterial and yeast systems – and yet show equivalent affinity to their targets compared to traditional antibodies.
The synthetic nanobodies used in this study came from three libraries designed recently by these scientists, which are based on the structures of nanobody-target complexes. Such sybodies – as they are called – can be modified to fit a given target protein within 12 days, whereas natural nanobodies take more than two months to be generated by repeated immunization and subsequent binder selection.
In the current study, planned and predefined conditions were provided for proper sybody selection, so that they actually recognize the prefusion spike conformation. This ability to choose the conditions for selection is also a big advantage of this technology. And lastly, nanobody formulations can be given by inhalation, which means that this could be developed into a convenient, rapid and direct mode of prevention.
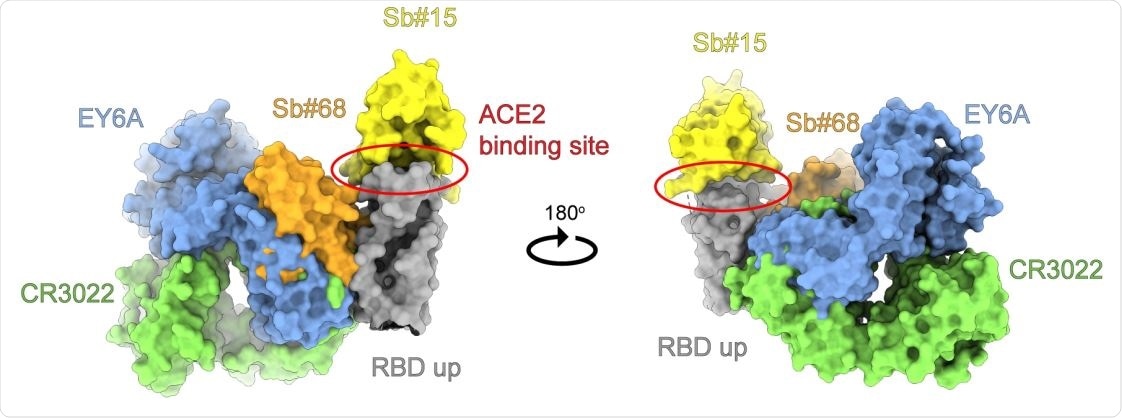
Anbody and sybody binding to a conserved RBD epitope. Superimposion of the crystal structures of the human crossreacve an-SARS-CoV-1 Fab fragment CR3022 (PDB ID: 6W41) and the human Fab fragment EY6A (PDB ID: 6ZCZ), both determined in complex with SARS-CoV-2 RBD, with homology models of sybodies Sb#15 and Sb#68 placed into the cryo-EM map of the symmetric 3up spike conformaon. Structures are shown as surface and colored as indicated.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Sybody generation and characterization
The researchers identified a set of 63 sybodies that bind two distinct epitopes at the RBD. There was enormous diversity in the binders generated during the search by three different institutes. These were then screened for ease of characterization, based on their biochemical features. Finally, of the 57 sybodies suitable for characterization, they found six sybodies with strong affinity for the RBD.
However, almost all the 57 sybodies effectively inhibited RBD's binding to the angiotensin-converting enzyme 2 (ACE2) receptor. But while most of them achieved a 50% reduction in RBD binding, five almost completely neutralized binding. These were part of the six mentioned earlier and "were able almost entirely to abolish the interaction between RBD and hACE2."
Simultaneous binding of sybody pairs to RBD
The six promising binders were Sb#14, Sb#15, Sb#16, Sb#42, Sb#45, and Sb#68. However, they found that when incubated with Sb#68, the spike protein could still bind powerfully to Sb#15. This could not occur if it was preincubated with any of the others.
The researchers considered this to be evidence that both Sb#68 and Sb#15 bind to the spike simultaneously. This was confirmed by additional studies using S-2P and S-6P, respectively, which are highly stabilized forms of the prefusion spike. When the full-length spike was tested, the two sybodies were known to bind cooperatively, unlike the purified RBD.
Moreover, they found that when these two sybodies were injected together, ACE2 binding was strongly inhibited by competitive binding. In fact, Sb#68 has over a ten-fold stronger affinity for the stabilized S-2P relative to the RBD alone.
Sb#14 and Sb#15 were found to inhibit binding to the viral receptor efficiently.
Neutralization of SARS-CoV-2
Individually, only the high-affinity sybodies (Sb#14 and Sb#15) showed high neutralization of pseudotyped SARS-CoV-2. When fused to human IgG1 Fc domains, the constructs showed much greater avidity, probably due to the bivalent presentation of the sybodies on the Fc domains. The most significant gain was with Sb#16, while it was small with Sb#68, indicating an epitope-dependent effect.
When both Sb#15 and Sb#68 were used in a pair, they showed potent neutralizing ability. This indicates their potential additive or synergistic neutralizing activity.
The pair of sybodies, Sb#15 and Sb#68 showed the ability to powerfully neutralize the entry of both pseudotyped and wildtype SARS-CoV-2, though the latter was less marked.
Bispecific sybody
The researchers then constructed three bodies, made up of Sb#15 and Sb#68 linked by a flexible GGGGS linker sequence with varying numbers of repetitions, from 2 to 6, called GS2, GS4 and GS6. They found that these bispecific sybodies had much higher affinity of binding over the individual sybodies.
They concluded, "The two sybodies of the fused construct bind simultaneously to the spike protein, thereby resulting in a strong avidity effect."
Their neutralizing effect was also much higher, by about a hundred-fold compared to the individual sybodies.
Binding study of fused sybodies by Cryo-EM
The researchers then examined the structure of the spike- Sb#15, spike- Sb#68, or spike-bispecific sybody complexes separately, by cryo-EM. The former two were at low resolution but served to assign the binding epitopes' general location on the RBD. Sb#15 and Sb#68 were seen to bind to the top of the RBD, overlapping strongly with the ACE2 binding site, and the side of the RBD, to a conserved and distinct epitope that is buried in the 'down' conformation. The Sb#68 epitope caused steric hindrance to ACE2 binding and this may be why this sybody can competitively inhibit ACE2.
However, the latter was seen in two different conformations. In the first, shown by about a third of the particles, the symmetry was threefold. The three RBDs were in the 'up' conformation, and each bound two sybodies, showing the simultaneous binding of Sb#15 and Sb#68.
In the second, shown by a fifth of the particles, the trimeric spike conformation was unusual, being asymmetric, with one RBD being in the 'up,' one in the 'up-out' and one in the 'down' position. In this scenario, the spike was bound to three Sb#15 and two Sb#68.
Of these, Sb#15 and Sb#68 were bound to one 'up' RBD as in the first conformation. The 'down' RBD bound only Sb#15, but this pushed the third one up and out. Thus, this prevented the threefold symmetry axis. The third 'up-out' RBD bound both sybodies, but Sb#68 showed weak density.
The same conformation was also present with the spike/Sb#15 complex, which confirms that this is responsible for the lack of the threefold symmetry axis. Thus, the 3-up state can be achieved only if both sybodies are bound in synergy.
The Sb#68/spike complex showed two 'up' RBDs bound to the sybody, but the third RBD showed weak density. Perhaps this was because it is intrinsically very flexible, which makes it hard to visualize. If in the one RBD 'up' and two RBDs' down', Sb#68 lacks access to the epitope because of steric hindrance by the neighboring RBD.
The mechanism by which the fused sybodies cause neutralization is not clear. It may be by receptor mimicry, whereby binding by sybodies causes the spike to shift to the post-fusion conformation before it binds to the ACE2 receptor. Since this is an irreversible event, it hinders viral entry.
Another possibility is that the sybodies just speed up spontaneous spike inactivation processes. Again, given that this pair of sybodies binds to two distinct epitopes, it may not be susceptible to escape mutations.
The many advantages of sybodies thus make this 'a robust platform to generate highly potent multi-specific biomolecules against coronaviruses." The rapid nature of sybody selection, development, and characterization demonstrate this platform's potential for fast-track responses to future pandemics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources