The transmissibility of the novel coronavirus, called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is high, while most infections appear to be very mild or asymptomatic. This has led to a pandemic of coronavirus disease 2019 (COVID-19) that has currently claimed over 1.96 million lives within one year. Scientists have come up with many vaccines at varying points of development. However, new variants of the virus continue to emerge, some of which are alleged to be even more transmissible or may possibly evade neutralization by the antibodies induced by newly approved vaccines.
One such strain is the South African 501.V2 and UK B1.1.7 strains, which have been isolated from many countries and seem to spread faster than the ancestral strain. A preprint that recently appeared on the bioRxiv* server reports the computational structure of this strain and the possible effect of the mutations on infectivity and neutralization.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The aim of the study
SARS-CoV-2 depends on its trimeric spike glycoprotein to achieve infection. The spike monomer is composed of two subunits, S1 and S2, with the receptor-binding domain (RBD) being located on the former. This region recognizes and binds to the host cell receptor, the angiotensin-converting enzyme 2 (ACE2). The S2 subunit mediates cell fusion and the internalization of the virus, after two cleavage steps catalyzed by host proteases, which cause a substantial change in protein conformation.
Most monoclonal antibodies and vaccines against SARS-CoV-2 at present are directed against the spike RBD. However, the UK variant has eight mutations in the spike, while the 501.V2 has three, both including N501Y in the RBD. The current study aims to understand the effect of the three mutations in the latter strain on RBD-ACE2 binding and neutralization by monoclonal antibodies targeting the spike. The researchers used molecular modeling and simulations to understand the structure of the mutant spike protein.
N501Y increases ACE2 binding affinity
The researchers found that the N501 residue forms two hydrogen bonds with two ACE2 residues that stabilize the RBD-ACE2 complex. When mutated to 501Y, three additional bonds are formed with residues like Y41 and K353, which increase the affinity. These residues have been established to be crucial to ACE2 binding with the SARS-CoV spike. Thus, the mutation is expected to enhance ACE2 affinity and thus promote transmission.
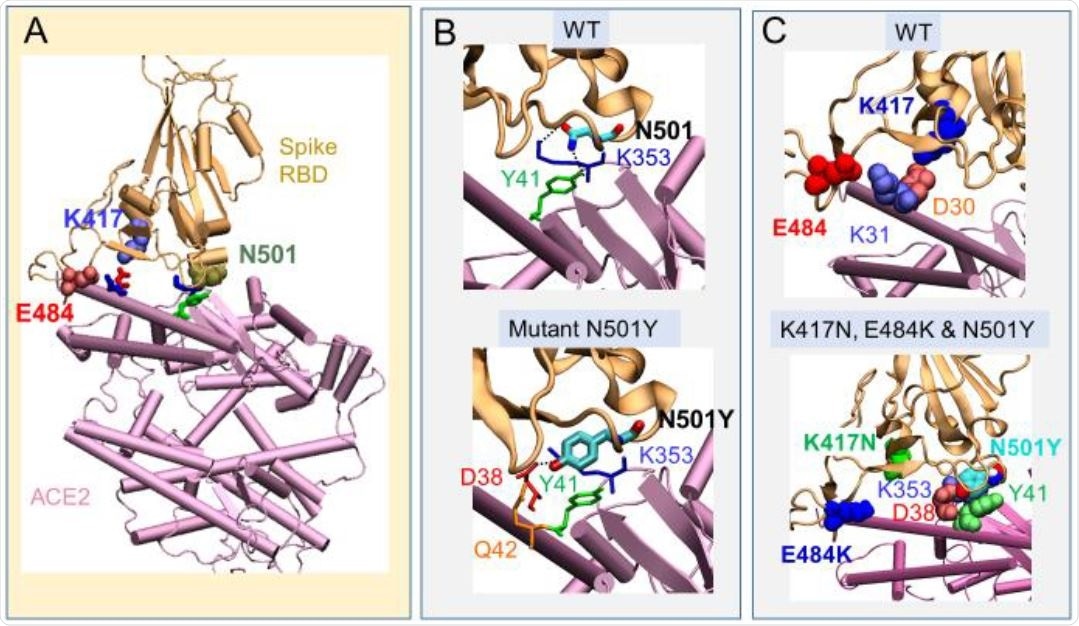
Change in interactions with ACE2 between wt RBD and Southern African variant 501.V2. (A) position of the three amino acids (K417, N501 and E584) at the interface with human ACE2 in the WT RBD; (B) comparison of the interactions of N501 (top) with those of the variant with single substitution N501Y (bottom). The latter enables a tighter contact result-ing in higher binding affinity compared to WT. (C) Same as B for the triple mutant K417N, D501Y and E484K. Two salt bridges originally present in the wt (top) are broken (bottom), suggesting a compensating effect by the substitutions E484K and K417N, countering N501Y.
K417N and E484K reduce binding affinity
However, the accompanying K417N and E484K mutations in the South African strain provide a counterpoise to the spike's increased affinity due to the N501Y mutation. They prevent the formation of two salt bridges that help to form and stabilize the RBD-ACE2 complex. This reduces ACE2 binding affinity. Thus, this strain is less infectious and less rapidly spreading than the UK strain. This is even though both share the latter substitution.
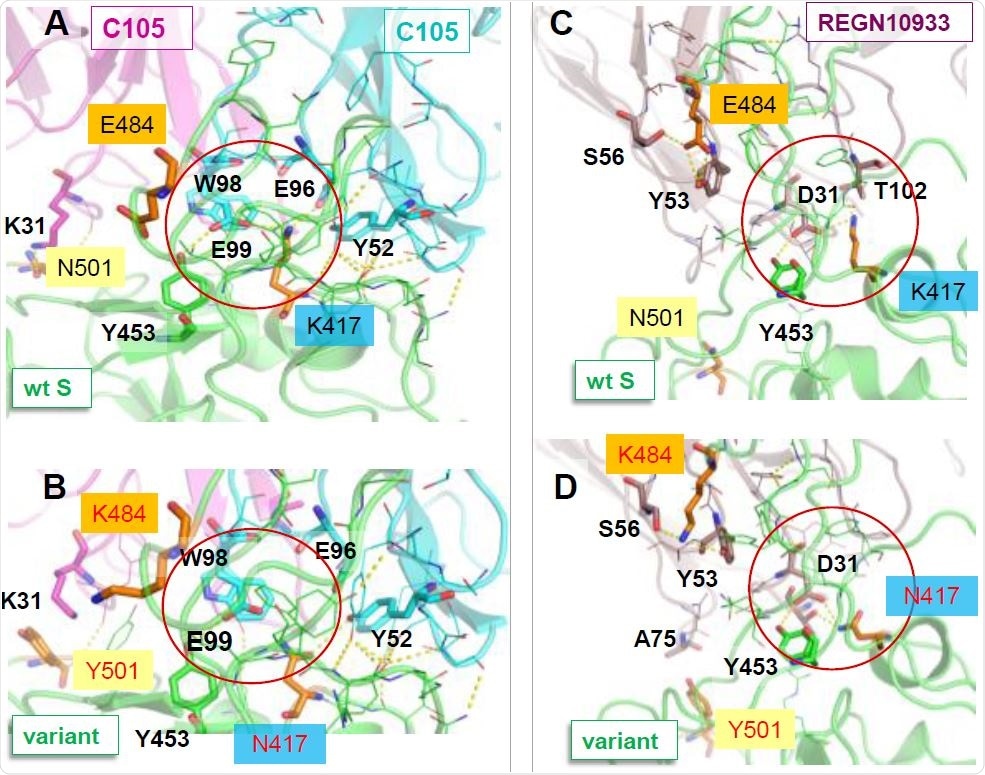
Disruption of salt bridges formed by K417 may weaken the association of C105 and REGN10933 with the South African var-iant 501.v2, compared to their interaction with the wt S. The panels compare the interactions of the mAbs C105 (A-B) and REGN10933 (C-D) with the wt RBD (A and C) and the South African mutant (B and D). A central salt bridge with C105 (K417-E99/E96) and another with REGN10933 (K417-D31) are lost due to the substitution K417N. A new cation-π interaction with K31 is formed upon N501Y mutation in D, which may compensate to restore the binding of REGN10933. N501Y makes no interfacial contacts, and E484K undergoes rear-rangements to alleviate the effect of charge change. The net effect due to those changes in interfacial interactions is a weakening in binding affinity by 0.4±0.2 kcal/mol.
Neutralizing antibodies mostly unaffected by triple mutation
Secondly, the researchers demonstrated that five of the eleven monoclonal antibodies that were structurally analyzed for binding to the mutant and ancestral strains do not have significant contacts at the mutation sites. Alternatively, they show additional interactions at such sites that compensate for any lack of affinity. This suggests that with most antibodies, including one of the two Regeneron monoclonals in their antibody cocktail, they continue to be at least as effective against the mutant strains as against the wildtype virus. With two others, namely, with the monoclonal antibodies B38 and 2-4, efficacy is increased up to twofold, as new favorable interactions occur between the residues.
With others like C105 and H11-14, the perturbed interactions at the binding interface due to the mutations K417N and/or E484K are too significant to be completely reversed by conformational rearrangement. For instance, with the former, the network of interactions that is formed via the K147 residue is abolished by the mutation at this site. However, new interactions is formed at this site and by 501Y, with other residues, but overall, binding affinity is reduced.
Weaker positive interactions, or even repulsions at some sites, are also observed with three other antibodies, including the other Regeneron antibody. This indicates that this variant could have a 2-4 weaker affinity for binding by these antibodies and thus escape neutralization.
The researchers also found that both N501 and K417 play an essential part in the dynamics of the RBD-ACE2 complex, as mechanical hinge centers or as regulatory sites.
What are the implications?
The study made use of already accumulated structural data on various ACE2-RBD or spike RBD-antibody complexes to predict the effects of various substitutions and deletions on binding affinity. They conclude, “Despite the stronger binding to ACE2 caused by the substitution N501Y, the South African 501.V2 variant that has undergone two additional mutations (K417N and E484K) is unlikely to exhibit an increased infectivity and likely transmissibility.”
When it comes to antibody escape, the effects were mixed. Either combinations of antibodies that target distinct non-overlapping epitopes or use different neutralization mechanisms should be developed to prevent mutational escape. In view of the availability of mutational maps, it is necessary to develop monoclonal antibodies that have binding sites with high resistance to viral escape. This could mean positioning epitopes outside the RBD or using epitopes shared by both SARS-CoV and SARS-CoV-2 since these do not typically tolerate mutation. The researchers say they recently identified such an antibody targeting the putative SARS-CoV-2 spike superantigenic-like motif. This is the antibody 6D3, which not only blocks the S1/S2 cleavage site, but also blocks the superantigenic site that may cause severe systemic inflammation in advanced COVID-19.
The study brings out the utility of computational predictions, which can be tested to understand the dose of an antibody required to overcome such mutations. With both therapeutic monoclonal antibodies and new viral variants being on the rise, “this type of in silico assisted genomic/molecular surveillance may provide feedback for accelerating the design of experimental studies in response to the pandemic.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.